In [1]:
%matplotlib inline
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
In [2]:
from sklearn.preprocessing import StandardScaler, MinMaxScaler
from sklearn.decomposition import PCA
from sklearn.manifold import TSNE
from sklearn.cluster import KMeans
from sklearn.metrics import adjusted_mutual_info_score
In [3]:
from sklearn.datasets import make_blobs
In [4]:
from umap import UMAP
In [5]:
plt.style.use('dark_background')
Clustering¶
Toy example to illustrate concepts¶
In [6]:
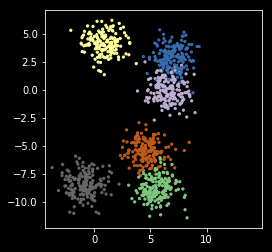
npts = 1000
nc = 6
X, y = make_blobs(n_samples=npts, centers=nc)
In [7]:
plt.scatter(X[:, 0], X[:, 1], s=5, c=y,
cmap=plt.cm.get_cmap('Accent', nc))
plt.axis('square')
pass
How to cluster¶
- Choose a clustering algorithm
- Choose the number of clusters
K-means¶
There are many different clustering methods, but K-means is fast, scales well, and can be interpreted as a probabilistic model. We will write 3 versions of the K-means algorithm to illustrate the main concepts. The algorithm is very simple:
- Start with \(k\) centers with labels \(0, 1, \ldots, k-1\)
- Find the distance of each data point to each center
- Assign the data points nearest to a center to its label
- Use the mean of the points assigned to a center as the new center
- Repeat for a fixed number of iterations or until the centers stop changing
Note: If you want a probabilistic interpretation, just treat the final solution as a mixture of (multivariate) normal distributions. K-means is often used to initialize the fitting of full statistical mixture models, which are computationally more demanding and hence slower.
Distance between sets of vectors¶
In [8]:
from scipy.spatial.distance import cdist
In [9]:
pts = np.arange(6).reshape(-1,2)
pts
Out[9]:
array([[0, 1],
[2, 3],
[4, 5]])
In [10]:
mus = np.arange(4).reshape(-1, 2)
mus
Out[10]:
array([[0, 1],
[2, 3]])
In [11]:
cdist(pts, mus)
Out[11]:
array([[0. , 2.82842712],
[2.82842712, 0. ],
[5.65685425, 2.82842712]])
Version 1¶
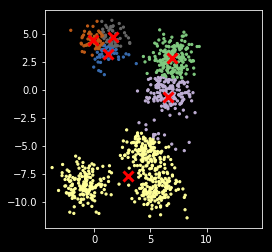
In [12]:
def my_kemans_1(X, k, iters=10):
"""K-means with fixed number of iterations."""
r, c = X.shape
centers = X[np.random.choice(r, k, replace=False)]
for i in range(iters):
m = cdist(X, centers)
z = np.argmin(m, axis=1)
centers = np.array([np.mean(X[z==i], axis=0) for i in range(k)])
return (z, centers)
In [13]:
np.random.seed(2018)
z, centers = my_kemans_1(X, nc)
Note that K-means can get stuck in local optimum¶
In [14]:
plt.scatter(X[:, 0], X[:, 1], s=5, c=z,
cmap=plt.cm.get_cmap('Accent', nc))
plt.scatter(centers[:, 0], centers[:, 1], marker='x',
linewidth=3, s=100, c='red')
plt.axis('square')
pass
Version 2¶
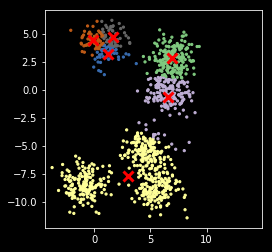
In [15]:
def my_kemans_2(X, k, tol= 1e-6):
"""K-means with tolerance."""
r, c = X.shape
centers = X[np.random.choice(r, k, replace=False)]
delta = np.infty
while delta > tol:
m = cdist(X, centers)
z = np.argmin(m, axis=1)
new_centers = np.array([np.mean(X[z==i], axis=0) for i in range(k)])
delta = np.sum((new_centers - centers)**2)
centers = new_centers
return (z, centers)
In [16]:
np.random.seed(2018)
z, centers = my_kemans_2(X, nc)
Still stuck in local optimum¶
In [17]:
plt.scatter(X[:, 0], X[:, 1], s=5, c=z,
cmap=plt.cm.get_cmap('Accent', nc))
plt.scatter(centers[:, 0], centers[:, 1], marker='x',
linewidth=3, s=100, c='red')
plt.axis('square')
pass
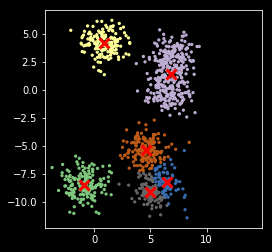
Version 3¶
Use of a score to evaluate the goodness of fit. In this case, the simplest score is just the sum of distances from each point to its nearest center.
In [18]:
def my_kemans_3(X, k, tol=1e-6, n_starts=10):
"""K-means with tolerance and random restarts."""
r, c = X.shape
best_score = np.infty
for i in range(n_starts):
centers = X[np.random.choice(r, k, replace=False)]
delta = np.infty
while delta > tol:
m = cdist(X, centers)
z = np.argmin(m, axis=1)
new_centers = np.array([np.mean(X[z==i], axis=0) for i in range(k)])
delta = np.sum((new_centers - centers)**2)
centers = new_centers
score = m[z].sum()
if score < best_score:
best_score = score
best_z = z
best_centers = centers
return (best_z, best_centers)
In [19]:
np.random.seed(2018)
z, centers = my_kemans_3(X, nc)
In [20]:
plt.scatter(X[:, 0], X[:, 1], s=5, c=z,
cmap=plt.cm.get_cmap('Accent', nc))
plt.scatter(centers[:, 0], centers[:, 1], marker='x',
linewidth=3, s=100, c='red')
plt.axis('square')
pass
Model selection¶
We use AMIS for model selection. You can also use likelihood-based
methods by interpreting the solution as a mixture of normals but we
don’t cover those approaches here. There are also other ad-hoc methods -
see sckikt-lean
docs
if you are interested.
Mutual Information Score (MIS)¶
The mutual information is defined as
It measures how much knowing \(X\) reduces the uncertainty about \(Y\) (and vice versa).
- If \(X\) is independent of \(Y\), then \(I(X; Y) = 0\)
- If \(X\) is a deterministic function of \(Y\), then \(I(X; Y) = 1\)
It is equivalent to the Kullback-Leibler divergence
We use AMIS (Adjusted MIS) here, which adjusts for the number of clusters used in the clustering method.
From the documentation
AMI(U, V) = [MI(U, V) - E(MI(U, V))] / [max(H(U), H(V)) - E(MI(U, V))]
In [21]:
ncols = 4
nrows = 2
plt.figure(figsize=(ncols*3, nrows*3))
for i, nc in enumerate(range(3, 11)):
kmeans = KMeans(nc, n_init=10)
clusters = kmeans.fit_predict(X)
centers = kmeans.cluster_centers_
amis = adjusted_mutual_info_score(y, clusters)
plt.subplot(nrows, ncols, i+1)
plt.scatter(X[:, 0], X[:, 1], s=5, c=y,
cmap=plt.cm.get_cmap('Accent', nc))
plt.scatter(centers[:, 0], centers[:, 1], marker='x',
linewidth=3, s=100, c='red')
plt.text(0.15, 0.9, 'nc = %d AMIS = %.2f' % (nc, amis),
fontsize=16, color='yellow',
transform=plt.gca().transAxes)
plt.xticks([])
plt.yticks([])
plt.axis('square')
plt.tight_layout()
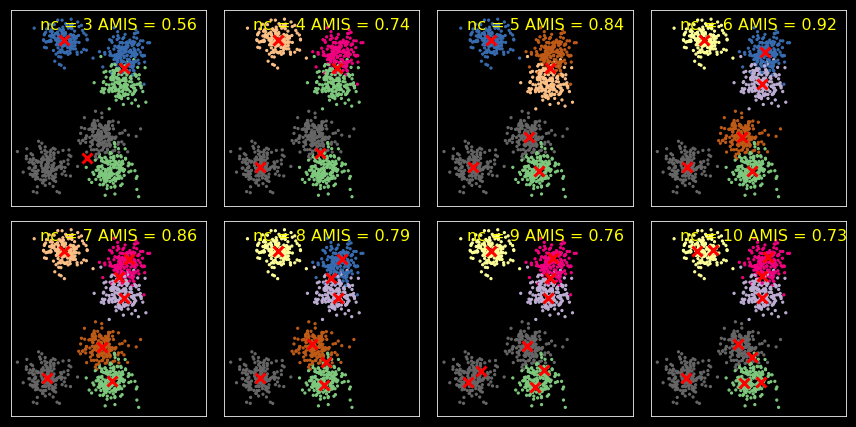
Comparing across samples¶
In [22]:
X1 = X + np.random.normal(0, 1, X.shape)
In [23]:
plt.figure(figsize=(8, 4))
for i, xs in enumerate([X, X1]):
plt.subplot(1,2,i+1)
plt.scatter(xs[:, 0], xs[:, 1], s=5, c=y,
cmap=plt.cm.get_cmap('Accent', nc))
plt.xticks([])
plt.yticks([])
plt.axis('square')
plt.tight_layout()
pass
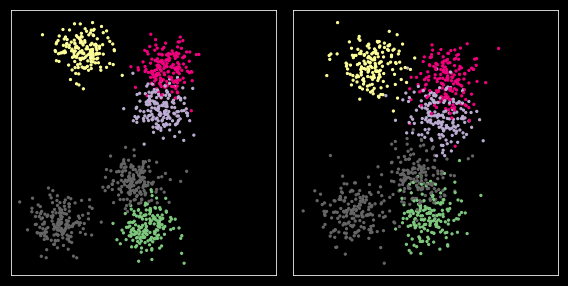
Option 1: Using reference sample¶
In [24]:
nc = 6
kmeans = KMeans(nc, n_init=10)
kmeans.fit(X)
z1 = kmeans.predict(X)
z2 = kmeans.predict(X1)
zs = [z1, z2]
In [25]:
plt.figure(figsize=(8, 4))
for i, xs in enumerate([X, X1]):
plt.subplot(1,2,i+1)
plt.scatter(xs[:, 0], xs[:, 1], s=5, c=zs[i],
cmap=plt.cm.get_cmap('Accent', nc))
plt.xticks([])
plt.yticks([])
plt.axis('square')
plt.tight_layout()
pass
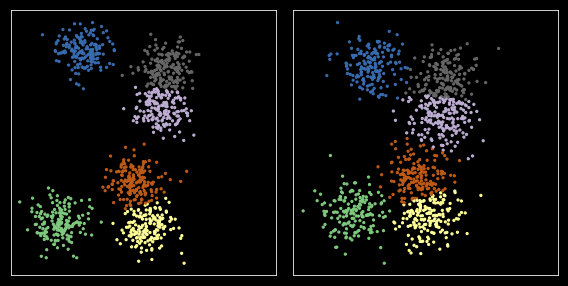
Option 2: Pooling¶
In [26]:
Y = np.r_[X, X1]
kmeans = KMeans(nc, n_init=10)
kmeans.fit(Y)
zs = np.split(kmeans.predict(Y), 2)
In [27]:
plt.figure(figsize=(8, 4))
for i, xs in enumerate([X, X1]):
plt.subplot(1,2,i+1)
plt.scatter(xs[:, 0], xs[:, 1], s=5, c=zs[i],
cmap=plt.cm.get_cmap('Accent', nc))
plt.xticks([])
plt.yticks([])
plt.axis('square')
plt.tight_layout()
pass
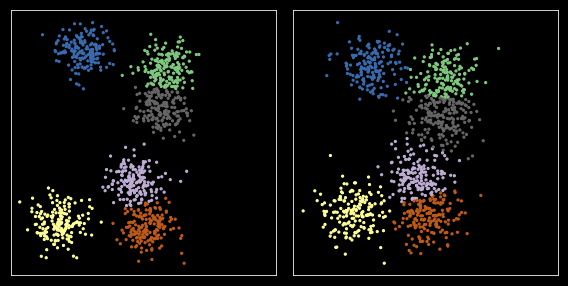
Option 3: Matching¶
In [28]:
from scipy.optimize import linear_sum_assignment
In [29]:
np.random.seed(2018)
kmeans = KMeans(nc, n_init=10)
c1 = kmeans.fit(X).cluster_centers_
z1 = kmeans.predict(X)
c2 = kmeans.fit(X1).cluster_centers_
z2 = kmeans.predict(X1)
zs = [z1, z2]
cs = [c1, c2]
In [30]:
plt.figure(figsize=(8, 4))
for i, xs in enumerate([X, X1]):
plt.subplot(1,2,i+1)
z = zs[i]
c = cs[i]
plt.scatter(xs[:, 0], xs[:, 1], s=5, c=z,
cmap=plt.cm.get_cmap('Accent', nc))
plt.xticks([])
plt.yticks([])
plt.axis('square')
plt.tight_layout()
pass
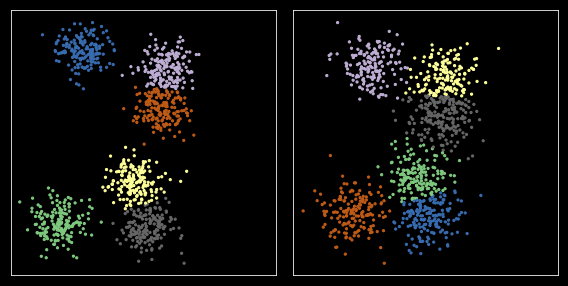
Define cost function¶
We use a simple cost as just the distance between centers. More complex dissimilarity measures could be used.
In [31]:
cost = cdist(c1, c2)
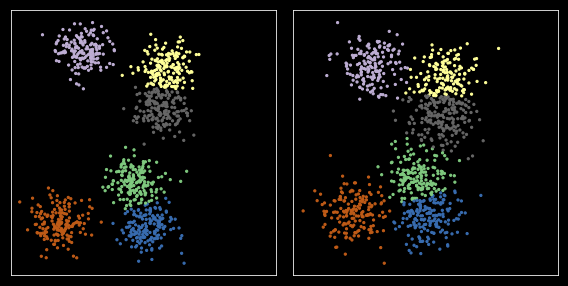
Run the Hungarian (Muunkres) algorithm for bipartitie matching¶
In [32]:
row_ind, col_ind = linear_sum_assignment(cost)
In [33]:
row_ind, col_ind
Out[33]:
(array([0, 1, 2, 3, 4, 5]), array([4, 2, 0, 1, 5, 3]))
Swap cluster indexes to align data sets¶
In [34]:
z1_aligned = col_ind[z1]
zs = [z1_aligned, z2]
In [35]:
plt.figure(figsize=(8, 4))
for i, xs in enumerate([X, X1]):
plt.subplot(1,2,i+1)
plt.scatter(xs[:, 0], xs[:, 1], s=5, c=zs[i],
cmap=plt.cm.get_cmap('Accent', nc))
plt.xticks([])
plt.yticks([])
plt.axis('square')
plt.tight_layout()
pass
In [ ]: