Capstone: Summarizing Clinical and Demographic Data¶
In [1]:
import numpy as np
import pandas as pd
from pandas import DataFrame, Series
In [2]:
pd.set_option('precision',2)
Load data sets from Excel¶
In [3]:
df_subject = pd.read_excel('data/capstone1/HIV Boot Camp.xls', sheetname='Patient Information')
df_hiv = pd.read_excel('data/capstone1/HIV Boot Camp.xls', sheetname='HIV RNA')
Queries on Patient Information table¶
In [4]:
df_subject.shape
Out[4]:
(3748, 10)
In [5]:
df_subject.head(n=10)
Out[5]:
| ID | AGE | RACE | SEX | MSM | IDU | UNC Entry to Care Year | Prior HIV Care Elsewhere | On ART | ART Type | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 38 | AMIND | F | No | No | 2012 | NO | Yes | Integrase |
| 1 | 2 | 44 | AMIND | F | No | No | 2006 | YES | No | NaN |
| 2 | 3 | 33 | AMIND | F | No | No | 2005 | NO | No | NaN |
| 3 | 4 | 60 | AMIND | F | No | Yes | 2003 | NO | Yes | Integrase |
| 4 | 6 | 47 | AMIND | F | No | No | 2001 | YES | Yes | Other |
| 5 | 7 | 54 | AMIND | F | No | No | 2000 | YES | Yes | Other |
| 6 | 8 | 46 | AMIND | F | No | No | 2000 | NO | No | NaN |
| 7 | 9 | 69 | AMIND | F | No | No | 1999 | NO | No | NaN |
| 8 | 10 | 41 | AMIND | F | No | No | 1999 | NO | No | NaN |
| 9 | 11 | 38 | AMIND | F | No | No | 1999 | NO | No | NaN |
In [6]:
df_subject.tail(n=5)
Out[6]:
| ID | AGE | RACE | SEX | MSM | IDU | UNC Entry to Care Year | Prior HIV Care Elsewhere | On ART | ART Type | |
|---|---|---|---|---|---|---|---|---|---|---|
| 3743 | 4253 | 44 | WHITE | M | No | No | 1990 | YES | No | NaN |
| 3744 | 4254 | 56 | WHITE | M | Yes | Yes | 1990 | YES | No | NaN |
| 3745 | 4255 | 58 | WHITE | M | Yes | No | 1990 | YES | Yes | Other |
| 3746 | 4257 | 60 | WHITE | M | Yes | No | 1989 | YES | Yes | Other |
| 3747 | 4259 | 57 | WHITE | M | Yes | No | 1985 | NO | Yes | Other |
In [7]:
df_subject.sample(n=8)
Out[7]:
| ID | AGE | RACE | SEX | MSM | IDU | UNC Entry to Care Year | Prior HIV Care Elsewhere | On ART | ART Type | |
|---|---|---|---|---|---|---|---|---|---|---|
| 595 | 662 | 52 | BLACK | F | No | Unknown | 1998 | NO | Yes | Integrase |
| 1509 | 1665 | 59 | BLACK | M | Yes | No | 2009 | NO | Yes | NNRTI |
| 3010 | 3436 | 62 | WHITE | M | Yes | Unknown | 2009 | NO | Yes | Integrase |
| 1143 | 1291 | 32 | BLACK | M | Yes | No | 2015 | NO | Yes | Integrase |
| 1190 | 1338 | 52 | BLACK | M | No | No | 2014 | NO | Yes | Integrase |
| 3657 | 4161 | 64 | WHITE | M | Yes | Unknown | 1994 | NO | No | NaN |
| 1772 | 1957 | 75 | BLACK | M | No | No | 2004 | NO | No | NaN |
| 3311 | 3752 | 71 | WHITE | M | Yes | No | 2001 | YES | Yes | PI |
Filtering on age¶
In [8]:
df_subject[df_subject.AGE >= 85]
Out[8]:
| ID | AGE | RACE | SEX | MSM | IDU | UNC Entry to Care Year | Prior HIV Care Elsewhere | On ART | ART Type | |
|---|---|---|---|---|---|---|---|---|---|---|
| 202 | 218 | 89 | BLACK | F | No | No | 2007 | YES | No | NaN |
| 703 | 800 | 89 | BLACK | F | No | No | 1995 | NO | Yes | Other |
| 736 | 840 | 88 | BLACK | F | No | No | 1994 | YES | No | NaN |
| 755 | 869 | 85 | BLACK | F | No | No | 1993 | YES | No | NaN |
| 917 | 1042 | 86 | WHITE | F | No | No | 2007 | YES | No | NaN |
| 1983 | 2191 | 85 | BLACK | M | No | No | 2001 | NO | No | NaN |
| 2297 | 2569 | 87 | BLACK | M | No | No | 1998 | YES | No | NaN |
| 3321 | 3762 | 88 | WHITE | M | Yes | No | 2001 | YES | No | NaN |
| 3606 | 4102 | 85 | WHITE | M | Unknown | Unknown | 1996 | YES | No | NaN |
| 3645 | 4149 | 86 | WHITE | M | No | No | 1995 | YES | Yes | Other |
Split-Apply-Combine¶
In [9]:
grouped = df_subject.groupby(['RACE', 'On ART'])
df1 = grouped[['ID']].count()
df1
Out[9]:
| ID | ||
|---|---|---|
| RACE | On ART | |
| AMIND | No | 39 |
| Yes | 24 | |
| ASIAN | No | 3 |
| Yes | 5 | |
| BLACK | No | 1164 |
| Yes | 1037 | |
| HISPN | No | 33 |
| Yes | 6 | |
| OTHER | No | 94 |
| Yes | 118 | |
| UNKNW | No | 5 |
| Yes | 16 | |
| WHITE | No | 601 |
| Yes | 603 |
Working with hierarchical indexes¶
In [10]:
df1.unstack(level = 1)
Out[10]:
| ID | ||
|---|---|---|
| On ART | No | Yes |
| RACE | ||
| AMIND | 39 | 24 |
| ASIAN | 3 | 5 |
| BLACK | 1164 | 1037 |
| HISPN | 33 | 6 |
| OTHER | 94 | 118 |
| UNKNW | 5 | 16 |
| WHITE | 601 | 603 |
In [11]:
df1.unstack(level=0)
Out[11]:
| ID | |||||||
|---|---|---|---|---|---|---|---|
| RACE | AMIND | ASIAN | BLACK | HISPN | OTHER | UNKNW | WHITE |
| On ART | |||||||
| No | 39 | 3 | 1164 | 33 | 94 | 5 | 601 |
| Yes | 24 | 5 | 1037 | 6 | 118 | 16 | 603 |
Perhaps surprising¶
In [12]:
df_subject.groupby(['SEX', 'MSM'])[['ID']].count()
Out[12]:
| ID | ||
|---|---|---|
| SEX | MSM | |
| F | No | 1071 |
| Unknown | 15 | |
| Yes | 3 | |
| M | No | 609 |
| Unknown | 467 | |
| Yes | 1583 |
Multiple summaries of single variable¶
In [13]:
grouped = df_subject.groupby(['RACE', 'SEX'])
df2 = grouped[['AGE']].agg(['count', 'mean', 'std'])
df2
Out[13]:
| AGE | ||||
|---|---|---|---|---|
| count | mean | std | ||
| RACE | SEX | |||
| AMIND | F | 19 | 45.26 | 8.25 |
| M | 44 | 50.34 | 9.41 | |
| ASIAN | F | 2 | 34.00 | 7.07 |
| M | 6 | 43.50 | 16.87 | |
| BLACK | F | 770 | 51.22 | 11.16 |
| M | 1431 | 49.13 | 12.78 | |
| HISPN | F | 11 | 46.09 | 7.67 |
| M | 28 | 49.04 | 10.26 | |
| OTHER | F | 51 | 46.57 | 11.47 |
| M | 161 | 46.20 | 11.21 | |
| UNKNW | F | 5 | 46.40 | 12.90 |
| M | 16 | 44.19 | 10.60 | |
| WHITE | F | 231 | 50.66 | 10.55 |
| M | 973 | 51.24 | 11.06 | |
In [14]:
df2.unstack(level=1)
Out[14]:
| AGE | ||||||
|---|---|---|---|---|---|---|
| count | mean | std | ||||
| SEX | F | M | F | M | F | M |
| RACE | ||||||
| AMIND | 19 | 44 | 45.26 | 50.34 | 8.25 | 9.41 |
| ASIAN | 2 | 6 | 34.00 | 43.50 | 7.07 | 16.87 |
| BLACK | 770 | 1431 | 51.22 | 49.13 | 11.16 | 12.78 |
| HISPN | 11 | 28 | 46.09 | 49.04 | 7.67 | 10.26 |
| OTHER | 51 | 161 | 46.57 | 46.20 | 11.47 | 11.21 |
| UNKNW | 5 | 16 | 46.40 | 44.19 | 12.90 | 10.60 |
| WHITE | 231 | 973 | 50.66 | 51.24 | 10.55 | 11.06 |
Extracting one sub-group¶
In [15]:
white_female = grouped.get_group(('WHITE', 'F'))
white_female.head()
Out[15]:
| ID | AGE | RACE | SEX | MSM | IDU | UNC Entry to Care Year | Prior HIV Care Elsewhere | On ART | ART Type | |
|---|---|---|---|---|---|---|---|---|---|---|
| 858 | 981 | 47 | WHITE | F | No | No | 2015 | NO | Yes | Integrase |
| 859 | 982 | 49 | WHITE | F | No | Unknown | 2015 | YES | Yes | Integrase |
| 860 | 983 | 59 | WHITE | F | No | No | 2015 | NO | Yes | Integrase |
| 861 | 984 | 51 | WHITE | F | No | No | 2015 | NO | Yes | Integrase |
| 862 | 985 | 57 | WHITE | F | No | Yes | 2015 | NO | Yes | NNRTI |
Merging data sets¶
In [16]:
df_hiv.head()
Out[16]:
| ID | Test Month | Test Year | HIV RNA | |
|---|---|---|---|---|
| 0 | 1 | 10 | 2011 | 64360 |
| 1 | 1 | 1 | 2012 | 17848 |
| 2 | 1 | 5 | 2012 | 28773 |
| 3 | 1 | 7 | 2012 | 23085 |
| 4 | 2 | 7 | 2005 | 5871 |
In [17]:
df_hiv.shape
Out[17]:
(12862, 4)
In [18]:
df_subject_hiv = pd.merge(df_subject, df_hiv, on='ID')
In [19]:
df_subject_hiv.shape
Out[19]:
(12877, 13)
In [20]:
df_subject_hiv.head()
Out[20]:
| ID | AGE | RACE | SEX | MSM | IDU | UNC Entry to Care Year | Prior HIV Care Elsewhere | On ART | ART Type | Test Month | Test Year | HIV RNA | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 38 | AMIND | F | No | No | 2012 | NO | Yes | Integrase | 10 | 2011 | 64360 |
| 1 | 1 | 38 | AMIND | F | No | No | 2012 | NO | Yes | Integrase | 1 | 2012 | 17848 |
| 2 | 1 | 38 | AMIND | F | No | No | 2012 | NO | Yes | Integrase | 5 | 2012 | 28773 |
| 3 | 1 | 38 | AMIND | F | No | No | 2012 | NO | Yes | Integrase | 7 | 2012 | 23085 |
| 4 | 2 | 44 | AMIND | F | No | No | 2006 | YES | No | NaN | 7 | 2005 | 5871 |
Viral load for one subject¶
In [21]:
df_subject_hiv[df_subject_hiv.ID == 1]
Out[21]:
| ID | AGE | RACE | SEX | MSM | IDU | UNC Entry to Care Year | Prior HIV Care Elsewhere | On ART | ART Type | Test Month | Test Year | HIV RNA | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 38 | AMIND | F | No | No | 2012 | NO | Yes | Integrase | 10 | 2011 | 64360 |
| 1 | 1 | 38 | AMIND | F | No | No | 2012 | NO | Yes | Integrase | 1 | 2012 | 17848 |
| 2 | 1 | 38 | AMIND | F | No | No | 2012 | NO | Yes | Integrase | 5 | 2012 | 28773 |
| 3 | 1 | 38 | AMIND | F | No | No | 2012 | NO | Yes | Integrase | 7 | 2012 | 23085 |
Who has the most time series data?¶
In [22]:
counts = df_subject_hiv.groupby(['ID'])[['HIV RNA']].count()
counts.head()
Out[22]:
| HIV RNA | |
|---|---|
| ID | |
| 1 | 4 |
| 2 | 6 |
| 3 | 3 |
| 4 | 2 |
| 6 | 3 |
In [23]:
subject_id = counts[counts['HIV RNA'] == counts['HIV RNA'].max()]
subject_id
Out[23]:
| HIV RNA | |
|---|---|
| ID | |
| 106 | 46 |
In [24]:
subject_id.index[0]
Out[24]:
106
In [25]:
(df_subject_hiv[df_subject_hiv.ID == subject_id.index[0]].
sort_values(['Test Year', 'Test Month']))
Out[25]:
| ID | AGE | RACE | SEX | MSM | IDU | UNC Entry to Care Year | Prior HIV Care Elsewhere | On ART | ART Type | Test Month | Test Year | HIV RNA | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 421 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 1 | 1998 | 11000 |
| 422 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 6 | 1998 | 9203 |
| 423 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 9 | 1998 | 7147 |
| 424 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 11 | 1998 | 7355 |
| 425 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 1 | 1999 | 12083 |
| 426 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 3 | 1999 | 400 |
| 427 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 8 | 1999 | 1800 |
| 428 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 8 | 1999 | 200000 |
| 429 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 10 | 1999 | 810 |
| 430 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 12 | 1999 | 10000 |
| 431 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 12 | 1999 | 10000 |
| 432 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 1 | 2000 | 13800 |
| 433 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 2 | 2000 | 20000 |
| 434 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 2 | 2001 | 4838 |
| 435 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 5 | 2001 | 4800 |
| 436 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 9 | 2001 | 16238 |
| 437 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 10 | 2001 | 10000 |
| 438 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 4 | 2002 | 19938 |
| 439 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 5 | 2002 | 19000 |
| 440 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 11 | 2002 | 33947 |
| 441 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 11 | 2002 | 30000 |
| 442 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 1 | 2003 | 30000 |
| 443 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 6 | 2003 | 10652 |
| 444 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 2 | 2004 | 24488 |
| 445 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 4 | 2004 | 400 |
| 446 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 10 | 2004 | 641 |
| 447 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 11 | 2004 | 600 |
| 448 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 12 | 2004 | 1914 |
| 449 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 3 | 2005 | 23588 |
| 450 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 3 | 2005 | 23000 |
| 451 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 6 | 2005 | 6577 |
| 452 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 8 | 2005 | 1000 |
| 453 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 12 | 2006 | 100000 |
| 454 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 12 | 2006 | 100000 |
| 455 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 2 | 2007 | 1058 |
| 456 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 10 | 2007 | 55 |
| 457 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 11 | 2007 | 55 |
| 458 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 7 | 2008 | 72 |
| 459 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 7 | 2008 | 72 |
| 460 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 7 | 2009 | 521 |
| 461 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 7 | 2009 | 400 |
| 462 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 5 | 2010 | 12556 |
| 463 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 5 | 2010 | 12550 |
| 464 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 5 | 2010 | 9561 |
| 465 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 5 | 2011 | 50 |
| 466 | 106 | 68 | BLACK | F | No | No | 2011 | YES | Yes | Other | 9 | 2011 | 49 |
Binning into age categories¶
In [26]:
df_subject_hiv['Age Grpup'] = pd.cut(df_subject_hiv['AGE'], [0, 30, 60, 105])
df_subject_hiv.groupby('Age Grpup')['HIV RNA'].agg(['mean', 'min', 'max'])
Out[26]:
| mean | min | max | |
|---|---|---|---|
| Age Grpup | |||
| (0, 30] | 168012 | 19 | 40306875 |
| (30, 60] | 99641 | 1 | 21923000 |
| (60, 105] | 115820 | 7 | 84545454 |
Exercises¶
1. Load the worksheet CD4 Count in the spreadsheet
data/HIV Boot Camp.xls into a data frame named df_cd4.
In [27]:
df_cd4 = pd.read_excel('data/HIV Boot Camp.xls', sheetname='CD4 Count')
2. How many rows and columns are there in df_cd4?
In [28]:
df_cd4.shape
Out[28]:
(3716, 4)
3. Show 6 random rows from df_cd4.
In [29]:
df_cd4.sample(6)
Out[29]:
| ID | Test Month | Test Year | CD4 Count | |
|---|---|---|---|---|
| 2671 | 3117 | 1 | 2004 | 473 |
| 1477 | 1650 | 5 | 2009 | 273 |
| 454 | 501 | 10 | 2000 | 523 |
| 2310 | 2627 | 8 | 1997 | 143 |
| 3574 | 4102 | 1 | 1996 | 10 |
| 1426 | 1597 | 1 | 2010 | 154 |
4. What are the largest and smallest CD4 counts?
In [30]:
col = 'CD4 Count'
df_cd4[col].min(), df_cd4[col].max()
Out[30]:
(0, 3900)
5. What were the largest and smallest CD4 counts for tests in Dec 2010?
In [31]:
df1 = df_cd4[(df_cd4['Test Month'] == 12) & (df_hiv['Test Year']==2010)]
df1[col].min(), df1[col].max()
Out[31]:
(9, 900)
In [32]:
df2 = df_cd4.groupby(['Test Year', 'Test Month']).get_group((2010, 12))
df2[col].min(), df2[col].max()
Out[32]:
(165, 782)
6. Sort df_cd4 first by ID (ascending), then by Test Year (descending), then by Test Month (ascending). Show the first 6 rows before and after sorting.
In [33]:
df_cd4.head()
Out[33]:
| ID | Test Month | Test Year | CD4 Count | |
|---|---|---|---|---|
| 0 | 1 | 1 | 2012 | 211 |
| 1 | 2 | 7 | 2005 | 388 |
| 2 | 3 | 3 | 2005 | 371 |
| 3 | 4 | 7 | 2003 | 392 |
| 4 | 6 | 6 | 2001 | 35 |
In [34]:
df_cd4.sort_values(['ID', 'Test Year', 'Test Month'], ascending=[True, False, True]).head()
Out[34]:
| ID | Test Month | Test Year | CD4 Count | |
|---|---|---|---|---|
| 0 | 1 | 1 | 2012 | 211 |
| 1 | 2 | 7 | 2005 | 388 |
| 2 | 3 | 3 | 2005 | 371 |
| 3 | 4 | 7 | 2003 | 392 |
| 4 | 6 | 6 | 2001 | 35 |
7. Create a new column CD4 Group that categorizes CD4 counts
into the ranges (0-199, 200-500, 500-4000).
In [35]:
df_cd4['CD4 Group'] = pd.cut(df_cd4[col], bins=[0,200,500,4000], right=False)
df_cd4.head()
Out[35]:
| ID | Test Month | Test Year | CD4 Count | CD4 Group | |
|---|---|---|---|---|---|
| 0 | 1 | 1 | 2012 | 211 | [200, 500) |
| 1 | 2 | 7 | 2005 | 388 | [200, 500) |
| 2 | 3 | 3 | 2005 | 371 | [200, 500) |
| 3 | 4 | 7 | 2003 | 392 | [200, 500) |
| 4 | 6 | 6 | 2001 | 35 | [0, 200) |
8. Find the mean and median CD4 count for each subject. Show these values for subjects with IDs in (10, 25, 30, 40, 50, 60).
In [36]:
grouped = df_cd4.groupby('ID')
summary = grouped[col].agg(['mean', 'median'])
summary.ix[[10,25,30,40,50,60]]
Out[36]:
| mean | median | |
|---|---|---|
| ID | ||
| 10 | 798 | 798 |
| 25 | 283 | 283 |
| 30 | 876 | 876 |
| 40 | 1117 | 1117 |
| 50 | 510 | 510 |
| 60 | 763 | 763 |
9. Merge df_subject with df_cd4 by joining on ID to
create a new data frame df_subject_cd4. Show the first 6 rows.
In [37]:
df_subject_cd4 = pd.merge(df_subject, df_cd4, on='ID')
df_subject_cd4.head()
Out[37]:
| ID | AGE | RACE | SEX | MSM | IDU | UNC Entry to Care Year | Prior HIV Care Elsewhere | On ART | ART Type | Test Month | Test Year | CD4 Count | CD4 Group | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 38 | AMIND | F | No | No | 2012 | NO | Yes | Integrase | 1 | 2012 | 211 | [200, 500) |
| 1 | 2 | 44 | AMIND | F | No | No | 2006 | YES | No | NaN | 7 | 2005 | 388 | [200, 500) |
| 2 | 3 | 33 | AMIND | F | No | No | 2005 | NO | No | NaN | 3 | 2005 | 371 | [200, 500) |
| 3 | 4 | 60 | AMIND | F | No | Yes | 2003 | NO | Yes | Integrase | 7 | 2003 | 392 | [200, 500) |
| 4 | 6 | 47 | AMIND | F | No | No | 2001 | YES | Yes | Other | 6 | 2001 | 35 | [0, 200) |
10. Find the mean, minimum and maximum CD4 count for subgroups differing in RACE AND SEX.
In [38]:
grouped = df_subject_cd4.groupby(['RACE', 'SEX'])
grouped[col].agg(['mean', 'min', 'max'])
Out[38]:
| mean | min | max | ||
|---|---|---|---|---|
| RACE | SEX | |||
| AMIND | F | 298.53 | 10 | 798 |
| M | 361.50 | 7 | 871 | |
| ASIAN | F | 331.50 | 283 | 380 |
| M | 326.33 | 4 | 688 | |
| BLACK | F | 372.72 | 3 | 3900 |
| M | 362.36 | 0 | 2000 | |
| HISPN | F | 359.82 | 29 | 812 |
| M | 373.33 | 14 | 1146 | |
| OTHER | F | 372.04 | 4 | 1062 |
| M | 361.09 | 7 | 1434 | |
| UNKNW | F | 473.80 | 296 | 660 |
| M | 448.69 | 14 | 848 | |
| WHITE | F | 349.27 | 9 | 1559 |
| M | 365.61 | 1 | 1817 |
Sneak Preview: Visualizing data¶
In [39]:
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib inline
In [40]:
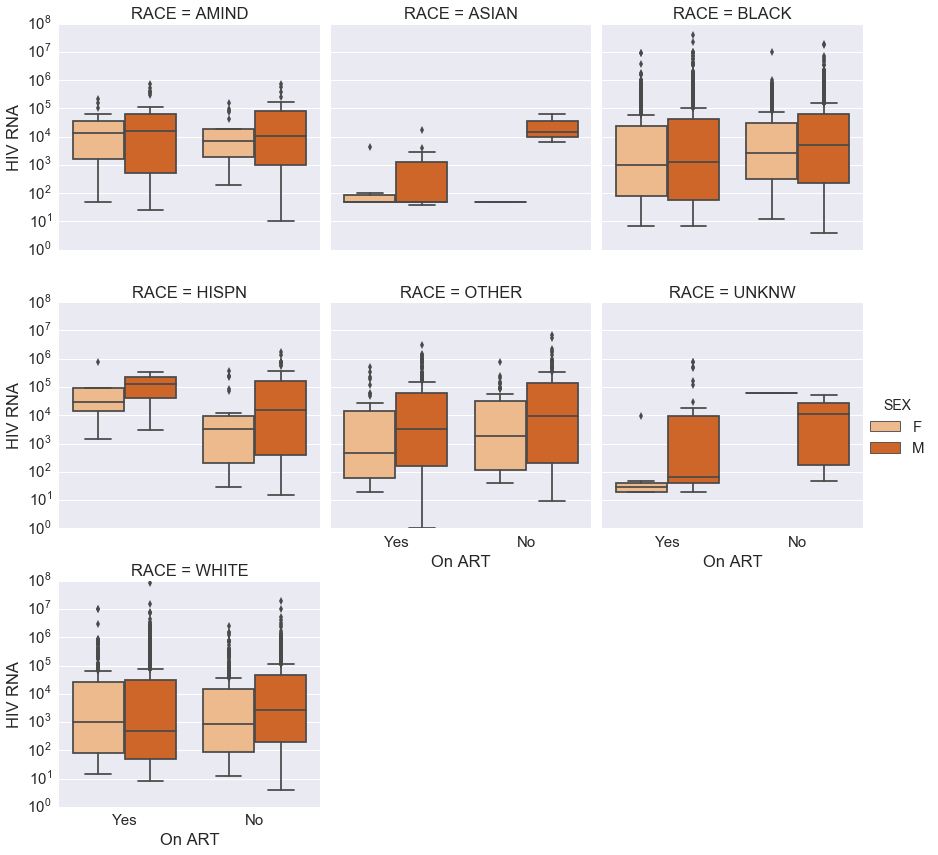
g = sns.factorplot(data=df_subject_hiv, x='On ART', y='HIV RNA',
col='RACE', col_wrap=3, kind='box', hue='SEX',
palette='Oranges')
g.axes[0].set_yscale('log')
pass