Numerical Evaluation of Integrals¶
Integration problems are common in statistics whenever we are dealing with continuous distributions. For example the expectation of a function is an integration problem
In Bayesian statistics, we need to solve the integration problem for the marginal likelihood or evidence
where \(\alpha\) is a hyperparameter and \(p(X \mid \alpha)\) appears in the denominator of Bayes theorem
In general, there is no closed form solution to these integrals, and we have to approximate them numerically. The first step is to check if there is some reparameterization that will simplify the problem. Then, the general approaches to solving integration problems are
- Numerical quadrature
- Importance sampling, adaptive importance sampling and variance reduction techniques (Monte Carlo swindles)
- Markov Chain Monte Carlo
- Asymptotic approximations (Laplace method and its modern version in variational inference)
This lecture will review the concepts for quadrature and Monte Carlo integration.
Quadrature¶
You may recall from Calculus that integrals can be numerically evaluated using quadrature methods such as Trapezoid and Simpson’s‘s rules. This is easy to do in Python, but has the drawback of the complexity growing as \(O(n^d)\) where \(d\) is the dimensionality of the data, and hence infeasible once \(d\) grows beyond a modest number.
Integrating functions¶
In [1]:
from scipy.integrate import quad
In [2]:
def f(x):
return x * np.cos(71*x) + np.sin(13*x)
In [3]:
x = np.linspace(0, 1, 100)
plt.plot(x, f(x))
pass
Exact solution¶
In [4]:
from sympy import sin, cos, symbols, integrate
x = symbols('x')
integrate(x * cos(71*x) + sin(13*x), (x, 0,1)).evalf(6)
Out[4]:
0.0202549
Multiple integration¶
Following the scipy.integrate
documentation,
we integrate
In [6]:
x, y = symbols('x y')
integrate(x*y, (x, 0, 1-2*y), (y, 0, 0.5))
Out[6]:
0.0104166666666667
In [7]:
from scipy.integrate import nquad
def f(x, y):
return x*y
def bounds_y():
return [0, 0.5]
def bounds_x(y):
return [0, 1-2*y]
y, err = nquad(f, [bounds_x, bounds_y])
y
Out[7]:
0.010416666666666668
Monte Carlo integration¶
The basic idea of Monte Carlo integration is very simple and only requires elementary statistics. Suppose we want to find the value of
this integral by estimating the fraction of random points that fall below \(f(x)\) multiplied by \(V\).
In a statistical context, we use Monte Carlo integration to estimate the expectation
with
We can estimate the Monte Carlo variance of the approximation as
Also, from the Central Limit Theorem,
The convergence of Monte Carlo integration is \(\mathcal{0}(n^{1/2})\) and independent of the dimensionality. Hence Monte Carlo integration generally beats numerical integration for moderate- and high-dimensional integration since numerical integration (quadrature) converges as \(\mathcal{0}(n^{d})\). Even for low dimensional problems, Monte Carlo integration may have an advantage when the volume to be integrated is concentrated in a very small region and we can use information from the distribution to draw samples more often in the region of importance.
An elementary, readable description of Monte Carlo integration and variance reduction techniques can be found here.
Intuition behind Monte Carlo integration¶
We want to find some integral
Consider the expectation of a function \(g(x)\) with respect to some distribution \(p(x)\). By defiitinon, we have
If we choose \(g(x) = f(x)/p(x)\), then we have
By the law of large numbers, the avaregae converges on the expectation, so we have
If \(f(x)\) is a proper integral (i.e. bounded), and \(p(x)\) is the uniform distribution, then \(g(x) = f(x)\) and this is known as ordinary Monte Carlo. If \(f(x)\) is imporper, then we need to use another distribtuion with the same support as \(f(x)\).
Intuition for error rate¶
We will just work this out for a proper integral \(f(x)\) defined in the unit cube and bounded by \(|f(x)| \le 1\). Draw a random uniform vector \(x\) in the unit cube. Then
Now consider summing over many such IID draws \(S_n = f(x_1) + f(x_2) + \cdots + f(x_n)\), :raw-latex:`\ldots`, x_n$. We have
and as expected, we see that \(I \approx S_n/n\). From Chebyshev’s inequality,
Suppose we want 1% accuracy and 99% confidence - i.e. set \(\epsilon = \delta = 0.01\). The above inequality tells us that we can achieve this with just \(n = 1/(\delta \epsilon^2) = 1,000,000\) samples, regardless of the data dimensionality.
Example¶
We want to estimate the following integral \(\int_0^1 e^x dx\). The minimum value of the function is 1 at \(x=0\) and \(e\) at \(x=1\).
In [1]:
x = np.linspace(0, 1, 100)
plt.plot(x, np.exp(x))
plt.xlim([0,1])
plt.ylim([0, np.e])
pass

Analytic solution¶
In [9]:
from sympy import symbols, integrate, exp
x = symbols('x')
expr = integrate(exp(x), (x,0,1))
expr.evalf()
Out[9]:
1.71828182845905
Using quadrature¶
In [10]:
from scipy import integrate
y, err = integrate.quad(exp, 0, 1)
y
Out[10]:
1.7182818284590453
Monte Carlo integration¶
In [2]:
for n in 10**np.array([1,2,3,4,5,6,7,8]):
x = np.random.uniform(0, 1, n)
sol = np.mean(np.exp(x))
print('%10d %.6f' % (n, sol))
10 1.748971
100 1.692790
1000 1.712451
10000 1.717981
100000 1.717453
1000000 1.718099
10000000 1.718495
100000000 1.718283
In [ ]:
#### Volumne of integration
Monitoring variance in Monte Carlo integration¶
We are often interested in knowing how many iterations it takes for Monte Carlo integration to “converge”. To do this, we would like some estimate of the variance, and it is useful to inspect such plots. One simple way to get confidence intervals for the plot of Monte Carlo estimate against number of iterations is simply to do many such simulations.
For the example, we will try to estimate the function (again)
In [12]:
def f(x):
return x * np.cos(71*x) + np.sin(13*x)
In [13]:
x = np.linspace(0, 1, 100)
plt.plot(x, f(x))
pass
Single MC integration estimate¶
In [14]:
n = 100
x = f(np.random.random(n))
y = 1.0/n * np.sum(x)
y
Out[14]:
0.13295443073705129
Using multiple independent sequences to monitor convergence¶
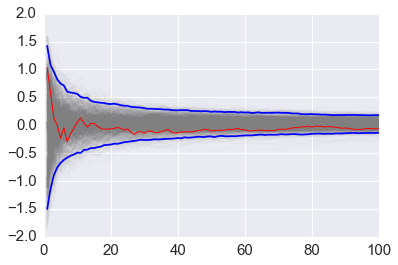
We vary the sample size from 1 to 100 and calculate the value of \(y = \sum{x}/n\) for 1000 replicates. We then plot the 2.5th and 97.5th percentile of the 1000 values of \(y\) to see how the variation in \(y\) changes with sample size. The blue lines indicate the 2.5th and 97.5th percentiles, and the red line a sample path.
In [15]:
n = 100
reps = 1000
x = f(np.random.random((n, reps)))
y = 1/np.arange(1, n+1)[:, None] * np.cumsum(x, axis=0)
upper, lower = np.percentile(y, [2.5, 97.5], axis=1)
In [16]:
plt.plot(np.arange(1, n+1), y, c='grey', alpha=0.02)
plt.plot(np.arange(1, n+1), y[:, 0], c='red', linewidth=1);
plt.plot(np.arange(1, n+1), upper, 'b', np.arange(1, n+1), lower, 'b')
pass
Using bootstrap to monitor convergence¶
If it is too expensive to do 1000 replicates, we can use a bootstrap instead.
In [17]:
xb = np.random.choice(x[:,0], (n, reps), replace=True)
yb = 1/np.arange(1, n+1)[:, None] * np.cumsum(xb, axis=0)
upper, lower = np.percentile(yb, [2.5, 97.5], axis=1)
In [18]:
plt.plot(np.arange(1, n+1)[:, None], yb, c='grey', alpha=0.02)
plt.plot(np.arange(1, n+1), yb[:, 0], c='red', linewidth=1)
plt.plot(np.arange(1, n+1), upper, 'b', np.arange(1, n+1), lower, 'b')
pass

Variance Reduction¶
With independent samples, the variance of the Monte Carlo estimate is
where \(Y_i = f(x_i)/p(x_i)\). The objective of Monte Carlo swindles is to make \(\text{Var}[\bar{g_n}]\) as small as possible for the same number of samples.
Change of variables¶
The Cauchy distribution is given by
Suppose we want to integrate the tail probability \(P(X > 3)\) using Monte Carlo. One way to do this is to draw many samples form a Cauchy distribution, and count how many of them are greater than 3, but this is extremely inefficient.
Only 10% of samples will be used¶
In [19]:
import scipy.stats as stats
h_true = 1 - stats.cauchy().cdf(3)
h_true
Out[19]:
0.10241638234956674
In [20]:
n = 100
x = stats.cauchy().rvs(n)
h_mc = 1.0/n * np.sum(x > 3)
h_mc, np.abs(h_mc - h_true)/h_true
Out[20]:
(0.14000000000000001, 0.36696880702301304)
A change of variables lets us use 100% of draws¶
We are trying to estimate the quantity
Using the substitution \(y = 3/x\) (and a little algebra), we get
Hence, a much more efficient MC estimator is
where \(y_i \sim \mathcal{U}(0, 1)\).
In [21]:
y = stats.uniform().rvs(n)
h_cv = 1.0/n * np.sum(3.0/(np.pi * (9 + y**2)))
h_cv, np.abs(h_cv - h_true)/h_true
Out[21]:
(0.10227041113815047, 0.0014252720909242859)
Monte Carlo swindles¶
Apart from change of variables, there are several general techniques for variance reduction, sometimes known as Monte Carlo swindles since these methods improve the accuracy and convergence rate of Monte Carlo integration without increasing the number of Monte Carlo samples. Some Monte Carlo swindles are:
- importance sampling
- stratified sampling
- control variates
- antithetic variates
- conditioning swindles including Rao-Blackwellization and independent variance decomposition
Most of these techniques are not particularly computational in nature, so we will not cover them in the course. I expect you will learn them elsewhere. We will illustrate importance sampling and antithetic variables here as examples.
Antithetic variables¶
The idea behind antithetic variables is to choose two sets of random numbers that are negatively correlated, then take their average, so that the total variance of the estimator is smaller than it would be with two sets of IID random variables.
In [22]:
def f(x):
return x * np.cos(71*x) + np.sin(13*x)
In [23]:
from sympy import sin, cos, symbols, integrate
x = symbols('x')
sol = integrate(x * cos(71*x) + sin(13*x), (x, 0,1)).evalf(16)
sol
Out[23]:
0.02025493910239406
Vanilla Monte Carlo¶
In [24]:
n = 10000
u = np.random.random(n)
x = f(u)
y = 1.0/n * np.sum(x)
y, abs(y-sol)/sol
Out[24]:
(0.0058850342599772341, 0.7094518907103678)
Antithetic variables use first half of u supplemented with 1-u¶
This works because the random draws are now negatively correlated, and hence the sum of the variances will be less than in the IID case, while the expectation is unchanged.
In [25]:
u = np.r_[u[:n//2], 1-u[:n//2]]
x = f(u)
y = 1.0/n * np.sum(x)
y, abs(y-sol)/sol
Out[25]:
(0.017795257018306126, 0.1214361628861775)
Importance sampling¶
Ordinary Monte Carlo sampling evaluates
Using another distribution \(h(x)\) - the so-called “importance function”, we can rewrite the above expression as an expectation with respect to \(h\)
giving us the new estimator
where \(x_i \sim g\) is a draw from the density \(h\). This is helpful if the distribution \(h\) has a similar shape as the function \(f(x)\) that we are integrating over, since we will draw more samples from places where the integrand makes a larger or more “important” contribution. This is very dependent on a good choice for the importance function \(h\). Two simple choices for \(h\) are scaling
and translation
In these cases, the parameter \(a\) is typically chosen using some adaptive algorithm, giving rise to adaptive importance sampling. Alternatively, a different distribution can be chosen as shown in the example below.
Example¶
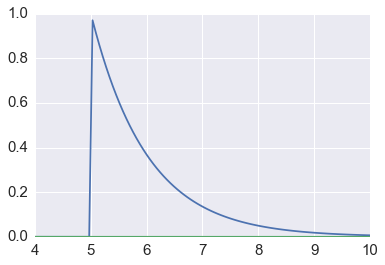
Suppose we want to estimate the tail probability of \(\mathcal{N}(0, 1)\) for \(P(X > 5)\). Regular MC integration using samples from \(\mathcal{N}(0, 1)\) is hopeless since nearly all samples will be rejected. However, we can use the exponential density truncated at 5 as the importance function and use importance sampling.
In [26]:
x = np.linspace(4, 10, 100)
plt.plot(x, stats.expon(5).pdf(x))
plt.plot(x, stats.norm().pdf(x))
pass
Expected answer¶
We expect about 3 draws out of 10,000,000 from \(\mathcal{N}(0, 1)\) to have a value greater than 5. Hence simply sampling from \(\mathcal{N}(0, 1)\) is hopelessly inefficient for Monte Carlo integration.
In [27]:
%precision 10
Out[27]:
'%.10f'
In [28]:
h_true =1 - stats.norm().cdf(5)
h_true
Out[28]:
0.0000002867
Using direct Monte Carlo integration¶
In [29]:
n = 10000
y = stats.norm().rvs(n)
h_mc = 1.0/n * np.sum(y > 5)
# estimate and relative error
h_mc, np.abs(h_mc - h_true)/h_true
Out[29]:
(0.0000000000, 1.0000000000)
Using importance sampling¶
In [30]:
n = 10000
y = stats.expon(loc=5).rvs(n)
h_is = 1.0/n * np.sum(stats.norm().pdf(y)/stats.expon(loc=5).pdf(y))
# estimate and relative error
h_is, np.abs(h_is- h_true)/h_true
Out[30]:
(0.0000002847, 0.0066615784)
Quasi-random numbers¶
Recall that the convergence of Monte Carlo integration is \(\mathcal{0}(n^{1/2})\). One issue with simple Monte Carlo is that randomly chosen points tend to be clumped. Clumping reduces accuracy since nearby points provide little additional information about the function begin estimated. One way to address this is to split the space into multiple integration regions, then sum them up. This is known as stratified sampling. Another alternative is to use quasi-random numbers which fill space more efficiently than random sequences
It turns out that if we use quasi-random or low discrepancy sequences, we can get convergence approaching \(\mathcal{0}(1/n)\). There are several such generators, but their use in statistical settings is limited to cases where we are integrating with respect to uniform distributions. The regularity can also give rise to errors when estimating integrals of periodic functions. However, these quasi-Monte Carlo methods are used in computational finance models.
Run
! pip install ghalton
if ghalton is not installed.
In [31]:
import ghalton
gen = ghalton.Halton(2)
In [32]:
plt.figure(figsize=(10,5))
plt.subplot(121)
xs = np.random.random((100,2))
plt.scatter(xs[:, 0], xs[:,1])
plt.axis([-0.05, 1.05, -0.05, 1.05])
plt.title('Pseudo-random', fontsize=20)
plt.subplot(122)
ys = np.array(gen.get(100))
plt.scatter(ys[:, 0], ys[:,1])
plt.axis([-0.05, 1.05, -0.05, 1.05])
plt.title('Quasi-random', fontsize=20);

In [33]:
h_true = 1 - stats.cauchy().cdf(3)
In [34]:
n = 10
x = stats.uniform().rvs((n, 5))
y = 3.0/(np.pi * (9 + x**2))
h_mc = np.sum(y, 0)/n
list(zip(h_mc, 100*np.abs(h_mc - h_true)/h_true))
Out[34]:
[(0.1046584475, 2.1891665182),
(0.1025211122, 0.1022588394),
(0.1041641176, 1.7064996957),
(0.1030694222, 0.6376322083),
(0.1016942178, 0.7051260477)]
In [35]:
gen1 = ghalton.Halton(1)
x = np.reshape(gen1.get(n*5), (n, 5))
y = 3.0/(np.pi * (9 + x**2))
h_qmc = np.sum(y, 0)/n
list(zip(h_qmc, 100*np.abs(h_qmc - h_true)/h_true))
Out[35]:
[(0.1026632536, 0.2410466633),
(0.1023042949, 0.1094428682),
(0.1026741252, 0.2516617574),
(0.1029118212, 0.4837496311),
(0.1026111501, 0.1901724534)]
In [ ]: