Statistics review 2: Samples and populations¶
R code accompanying paper
Key learning points¶
- A P value is the probability that an observed effect is simply due to chance; it therefore provides a measure of the strength of an association.
- P values are affected both by the magnitude of the effect and by the size of the study from which they are derived, and should therefore be interpreted with caution.
- If the purpose is Descriptive use standard Deviation; if the purpose is Estimation use standard Error
suppressPackageStartupMessages(library(tidyverse))
options(repr.plot.width=4, repr.plot.height=3)
Utility function from http://www.cookbook-r.com/Graphs/Multiple_graphs_on_one_page_(ggplot2)/
# Multiple plot function
#
# ggplot objects can be passed in ..., or to plotlist (as a list of ggplot objects)
# - cols: Number of columns in layout
# - layout: A matrix specifying the layout. If present, 'cols' is ignored.
#
# If the layout is something like matrix(c(1,2,3,3), nrow=2, byrow=TRUE),
# then plot 1 will go in the upper left, 2 will go in the upper right, and
# 3 will go all the way across the bottom.
#
multiplot <- function(..., plotlist=NULL, file, cols=1, layout=NULL) {
library(grid)
# Make a list from the ... arguments and plotlist
plots <- c(list(...), plotlist)
numPlots = length(plots)
# If layout is NULL, then use 'cols' to determine layout
if (is.null(layout)) {
# Make the panel
# ncol: Number of columns of plots
# nrow: Number of rows needed, calculated from # of cols
layout <- matrix(seq(1, cols * ceiling(numPlots/cols)),
ncol = cols, nrow = ceiling(numPlots/cols))
}
if (numPlots==1) {
print(plots[[1]])
} else {
# Set up the page
grid.newpage()
pushViewport(viewport(layout = grid.layout(nrow(layout), ncol(layout))))
# Make each plot, in the correct location
for (i in 1:numPlots) {
# Get the i,j matrix positions of the regions that contain this subplot
matchidx <- as.data.frame(which(layout == i, arr.ind = TRUE))
print(plots[[i]], vp = viewport(layout.pos.row = matchidx$row,
layout.pos.col = matchidx$col))
}
}
}
The Normal distribution¶
xp <- seq(-3, 3, length.out = 100)
y1 <- dnorm(xp, mean=0, sd=1)
y2 <- dnorm(xp, mean=0, sd=0.5)
y3 <- dnorm(xp, mean=0, sd=1.5)
df1 <- data.frame(x=xp, y1=y1, y2=y2, y3=y3)
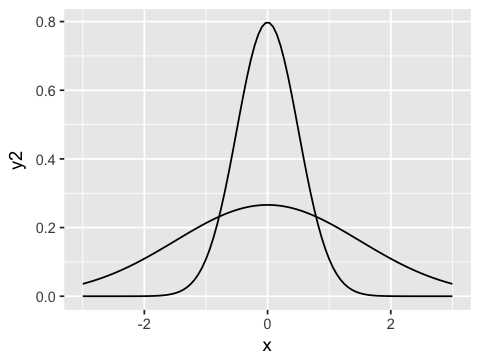
Small and large standard deviations¶
ggplot(df1, aes(x=x, y=y2)) + geom_line() + geom_line(aes(y=y3))
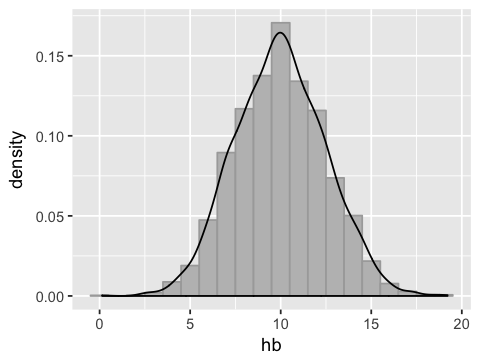
Normally distributed data¶
hb <- rnorm(2849, mean=10, sd=2.5)
df2 <- data.frame(hb = hb)
ggplot(df2, aes(x=hb)) +
geom_histogram(binwidth=1, fill='grey', color='darkgrey', aes(y=..density..)) +
geom_density()
What is the range of hb values that contains 95% of the mass?¶
mu = mean(df2$hb)
sigma = sd(df2$hb)
round(c(mu - 1.96 * sigma, mu + 1.96 * sigma), 2)
- 5.11
- 14.85
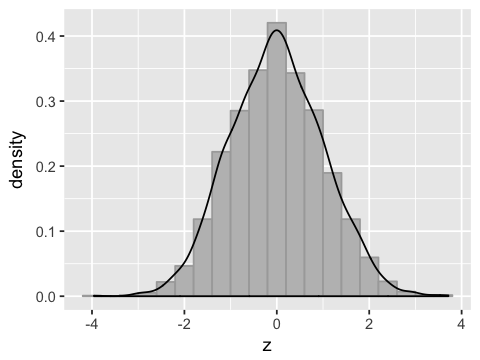
Converting to standard normal¶
df2$z <- (df2$hb - mu)/sigma
ggplot(df2, aes(x=z)) +
geom_histogram(binwidth=0.4, fill='grey', color='darkgrey', aes(y=..density..)) +
geom_density()
Area under normal curve¶
plot_area <- function(lower, upper) {
x <- seq(-3,3,length.out = 100)
y <- dnorm(x, 0, 1)
df <- data.frame(x=x, y=y)
area <- round(100 * pnorm(upper) - pnorm(lower), 2)
ggplot(df, aes(x=x, y=y)) + geom_line() +
stat_function(fun = dnorm,
xlim = c(lower, upper),
geom = "area",
fill='salmon') +
scale_x_continuous(breaks=c(lower, upper)) +
annotate("text", label = paste(area, "%", sep=""), size=3, x = 2, y = 0.3) +
xlab("") + ylab("") +
theme(axis.text.y = element_blank(),
axis.ticks.y = element_blank())
}
g1 <- plot_area(-1, 1)
g2 <- plot_area(-2, 2)
g3 <- plot_area(-3, 3)
g4 <- plot_area(-1.96, 1.96)
multiplot(g1, g2, g3, g4, cols = 2)
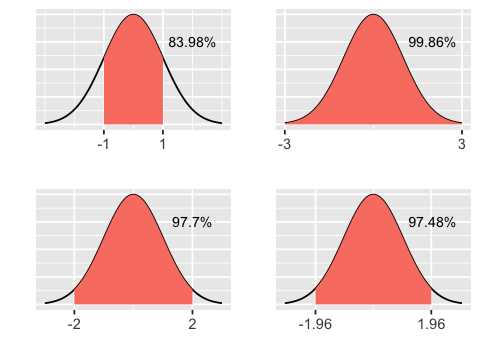
From sample to population¶
n.samples <- 100
n.reps <- 1000
x <- replicate(n.reps,runif(n.samples))
df3 <- data.frame(x=x[,1])
ggplot(df3, aes(x=x)) +
geom_histogram(bins=12, fill='grey', color='darkgrey', aes(y=..density..)) +
geom_density()
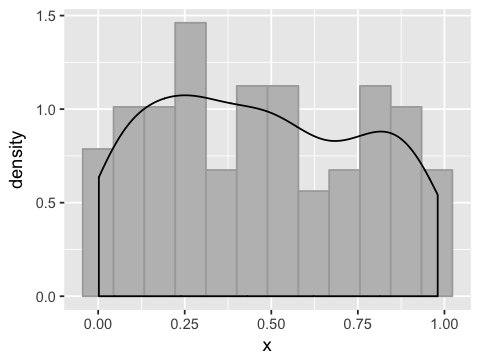
This is a consequence of the Central Limit Theorem - sums of independent random variables are normally distributed.
mus <- apply(x, 2, mean)
df4 <- data.frame(mus=mus)
ggplot(df4, aes(x=mus)) +
geom_histogram(bins=12, fill='grey', color='darkgrey', aes(y=..density..)) +
geom_density()
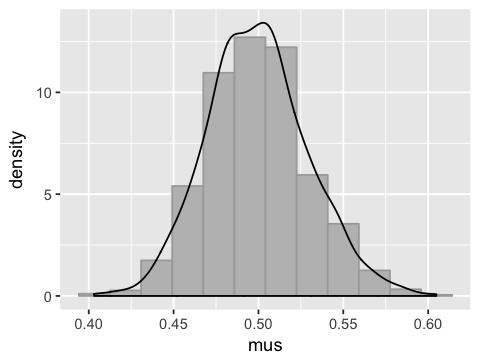
The standard deviation of the means (or standard error) is the sample standard deviation divided by the square root of the number of samples
round(sd(mus), 3)
round(sd(x[,1])/sqrt(length(x[,1])), 3)
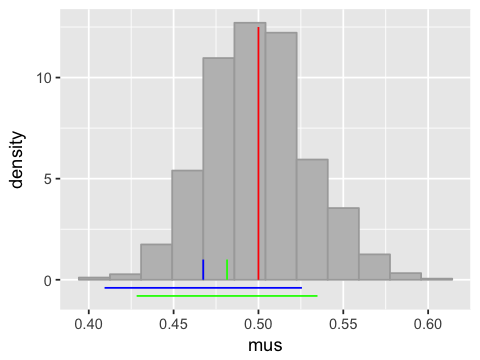
The standard error and confidence interval¶
Plot showing two 95% confidence intervals and the true population mean¶
# Standardize the sample means
z <- (mus - mean(mus))/sd(mus)
# Find 95% confidence intervals of two sample means
x1.lo <- mean(x[,1]) - 1.96*sd(x[,1])/sqrt(n.samples)
x1.hi <- mean(x[,1]) + 1.96*sd(x[,1])/sqrt(n.samples)
x2.lo <- mean(x[,2]) - 1.96*sd(x[,2])/sqrt(n.samples)
x2.hi <- mean(x[,2]) + 1.96*sd(x[,2])/sqrt(n.samples)
# 95% of the mass lies within +/- 1.96 of the SE
ggplot(df4, aes(x=mus)) +
geom_histogram(bins=12, fill='grey', color='darkgrey', aes(y=..density..)) +
annotate("segment", x = x1.lo, xend = x1.hi, y = -0.4, yend = -0.4, colour = "blue") +
annotate("segment", x = x2.lo, xend = x2.hi, y = -0.8, yend = -0.8, colour = "green") +
annotate("segment", x = mean(x[,1]), xend = mean(x[,1]), y = 0, yend = 1, colour = "blue") +
annotate("segment", x = mean(x[,2]), xend = mean(x[,2]), y = 0, yend = 1, colour = "green") +
annotate("segment", x = 0.5, xend = 0.5, y = 0, yend = 12.5, colour = "red")
Confidence intervals for smaller samples¶
x <- seq(-3, 3, length.out = 100)
y1 <- dnorm(x)
y2 <- dt(x, df=4)
y3 <- dt(x, df=19)
df5 <- data.frame(x=x, normal=y1, t4=y2, t19=y3)
ggplot(df5, aes(x=x, y=normal)) +
geom_line(color='blue') + # normal
geom_line(aes(y=t4), color='red') # T with 4 degrees of freedom
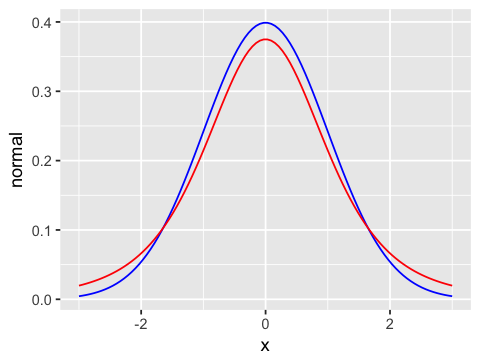
ggplot(df5, aes(x=x, y=normal)) +
geom_line(color='blue') + # normal
geom_line(aes(y=t19), color='red') # T with 19 degrees of freedom
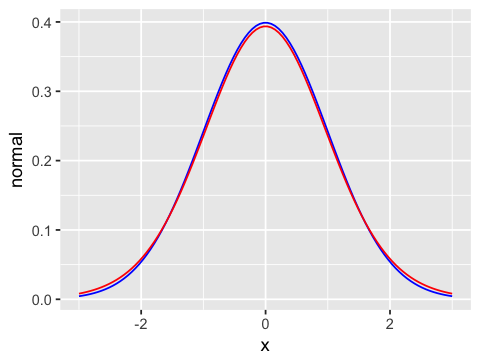
Scaling factors for confidence interval with diffent sample sizes¶
dfs <- c(10, 20, 30, 40, 50, 200)
k <- round(qt(0.975, df=dfs), 2)
df6 <- data.frame(n=dfs, k=k)
df6
| n | k |
|---|---|
| 10 | 2.23 |
| 20 | 2.09 |
| 30 | 2.04 |
| 40 | 2.02 |
| 50 | 2.01 |
| 200 | 1.97 |
Exercises¶
1. Load the file data/data.csv into a data frame. Make a
histogram of the column x data.
2. Calculate the mean and 90% reference range of x.
3. Calculate the standard error and 90% confidence intervals for the
estimated mean of x.
4. Write a function that takes a collection of numbers as input and returns the standard error.