Statistics review 13: Receiver operating characteristic
R code accompanying
paper
Key learning points
- Methods for assessing the performance of a diagnostic test
- Sensitivity, specificity and likelihood ratio of a test
- Receiver operating characteristic curve and the area under the curve
suppressPackageStartupMessages(library(tidyverse))
options(repr.plot.width=4, repr.plot.height=3)
Data
died <- c(81, 45)
survived <- c(591, 674)
df1 <- data.frame(died=died, survived=survived,
row.names=c("Lactate >1.5 mmol/l", "Lactate ≤1.5 mmol/l"))
df1
| died | survived |
|---|
| Lactate >1.5 mmol/l | 81 | 591 |
|---|
| Lactate ≤1.5 mmol/l | 45 | 674 |
|---|
Data set with different prevalence
died <- c(386, 214)
survived <- c(370, 421)
df2 <- data.frame(died=died, survived=survived,
row.names=c("Lactate >1.5 mmol/l", "Lactate ≤1.5 mmol/l"))
df2
| died | survived |
|---|
| Lactate >1.5 mmol/l | 386 | 370 |
|---|
| Lactate ≤1.5 mmol/l | 214 | 421 |
|---|
Sensitivity and specificity
prop.ci <- function(p, n, alpha=0.05) {
se <- sqrt(p*(1-p)/n)
k <- qnorm(1 - alpha/2)
round(c(p - k*se, p + k*se), 2)
}
sens <- function(tbl) {
a <- tbl[1,1]
b <- tbl[1,2]
c <- tbl[2,1]
d <- tbl[2,2]
n <- a + c
p <- a/n
round(p, 2)
}
0.64
p <- sens(df1)
n <- sum(df1)
prop.ci(p, n)
- 0.61
- 0.67
spec <- function(tbl) {
a <- tbl[1,1]
b <- tbl[1,2]
c <- tbl[2,1]
d <- tbl[2,2]
n <- b + d
p <- d/n
round(p, 2)
}
0.53
Positive and negative predictive values
ppv <- function(tbl) {
a <- tbl[1,1]
b <- tbl[1,2]
c <- tbl[2,1]
d <- tbl[2,2]
n <- a + b
p <- a/n
round(p, 2)
}
0.12
npv <- function(tbl, alpha=0.05) {
a <- tbl[1,1]
b <- tbl[1,2]
c <- tbl[2,1]
d <- tbl[2,2]
n <- c + d
p <- d/n
round(p, 2)
}
0.94
Effect of prevalence
c(sens(df1)[1], sens(df2)[1])
- 0.64
- 0.64
c(spec(df1)[1], spec(df2)[1])
- 0.53
- 0.53
c(ppv(df1)[1], ppv(df2)[1])
- 0.12
- 0.51
c(npv(df1)[1], npv(df2)[1])
- 0.94
- 0.66
Likelihood ratios
lr <- function(tbl) {
round(sens(tbl)[1]/(1 - spec(tbl)[1]), 2)
}
1.36
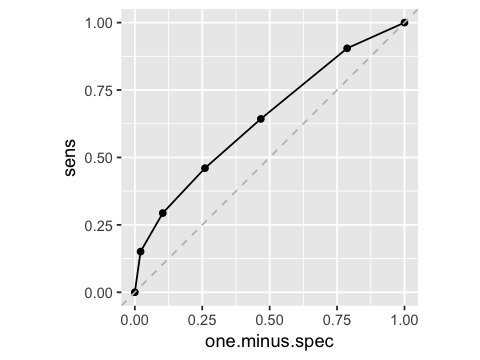
Receiver operating characteristic curve (ROC)
threshold <- c(0,1,1.5,2,3,5,25)
died <- c(126,114,81,58,37,19,0)
survived <- c(1265,996,591,329,131,27,0)
df3 <- data.frame(threshold=threshold, died=died, survived=survived)
df3
| threshold | died | survived |
|---|
| 0.0 | 126 | 1265 |
| 1.0 | 114 | 996 |
| 1.5 | 81 | 591 |
| 2.0 | 58 | 329 |
| 3.0 | 37 | 131 |
| 5.0 | 19 | 27 |
| 25.0 | 0 | 0 |
n.died <- 126
n.survived <- 1265
sens.1 <-function(died, ndied) {
a <- died
c <- ndied-died
a/(a + c)
}
spec.1 <-function(survived, nsurvived) {
b <- survived
d <- nsurvived - survived
b/(b+d)
}
df4 <- df3 %>% mutate(sens=sens.1(died, n.died), one.minus.spec=spec.1(survived, n.survived))
df4
| threshold | died | survived | sens | one.minus.spec |
|---|
| 0.0 | 126 | 1265 | 1.0000000 | 1.00000000 |
| 1.0 | 114 | 996 | 0.9047619 | 0.78735178 |
| 1.5 | 81 | 591 | 0.6428571 | 0.46719368 |
| 2.0 | 58 | 329 | 0.4603175 | 0.26007905 |
| 3.0 | 37 | 131 | 0.2936508 | 0.10355731 |
| 5.0 | 19 | 27 | 0.1507937 | 0.02134387 |
| 25.0 | 0 | 0 | 0.0000000 | 0.00000000 |
ggplot(df4, aes(x=one.minus.spec, y=sens)) + geom_point() + geom_line() +
geom_abline(intercept = 0, slope = 1, color='grey', linetype='dashed') +
coord_fixed()
Area under the curve (AUCROC)
suppressPackageStartupMessages(require(pracma))
round(-1 * trapz(x=df4$one.minus.spec, y=df4$sens), 2)
0.64
