Statistics review 14: Logistic regression
R code accompanying
paper
Key learning points
- Modeling the dependence of a binary response variable on one or more
explanatory variables
suppressPackageStartupMessages(library(tidyverse))
options(repr.plot.width=4, repr.plot.height=3)
marker <- seq(0.75, 4.75, by=0.5)
patients <- c(182, 233, 224, 236, 225, 215, 221, 200, 264)
deaths <- c(7, 27, 44, 91, 130, 168, 194, 191, 260)
df <- data.frame(marker=marker, patients=patients, deaths=deaths)
df
| marker | patients | deaths |
|---|
| 0.75 | 182 | 7 |
| 1.25 | 233 | 27 |
| 1.75 | 224 | 44 |
| 2.25 | 236 | 91 |
| 2.75 | 225 | 130 |
| 3.25 | 215 | 168 |
| 3.75 | 221 | 194 |
| 4.25 | 200 | 191 |
| 4.75 | 264 | 260 |
df1 <- df %>% mutate(prop=deaths/patients)
df1
| marker | patients | deaths | prop |
|---|
| 0.75 | 182 | 7 | 0.03846154 |
| 1.25 | 233 | 27 | 0.11587983 |
| 1.75 | 224 | 44 | 0.19642857 |
| 2.25 | 236 | 91 | 0.38559322 |
| 2.75 | 225 | 130 | 0.57777778 |
| 3.25 | 215 | 168 | 0.78139535 |
| 3.75 | 221 | 194 | 0.87782805 |
| 4.25 | 200 | 191 | 0.95500000 |
| 4.75 | 264 | 260 | 0.98484848 |
ggplot(df1, aes(x=marker, y=prop)) + geom_point()
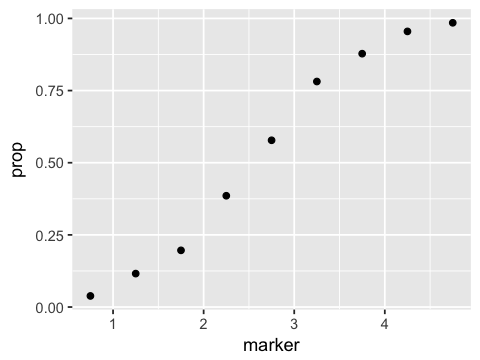
df2 <- df1 %>% mutate(logit=log(prop/(1-prop)))
df2
| marker | patients | deaths | prop | logit |
|---|
| 0.75 | 182 | 7 | 0.03846154 | -3.2188758 |
| 1.25 | 233 | 27 | 0.11587983 | -2.0320393 |
| 1.75 | 224 | 44 | 0.19642857 | -1.4087672 |
| 2.25 | 236 | 91 | 0.38559322 | -0.4658742 |
| 2.75 | 225 | 130 | 0.57777778 | 0.3136576 |
| 3.25 | 215 | 168 | 0.78139535 | 1.2738164 |
| 3.75 | 221 | 194 | 0.87782805 | 1.9720213 |
| 4.25 | 200 | 191 | 0.95500000 | 3.0550489 |
| 4.75 | 264 | 260 | 0.98484848 | 4.1743873 |
ggplot(df2, aes(x=marker, y=logit)) + geom_point()
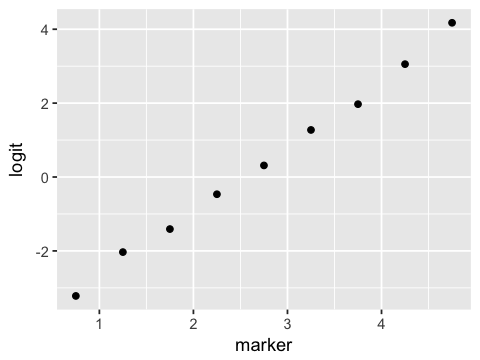
Maximum likelihood estimation
The parameters of many statistical models are estimated by maximizing
the likelihood. This likelihood is a function of the unnormalized
probability density after having observed some data. We will illustrate
by example to give some intuition.
Simulating sample data come from a population
mu <- 4
sigma <- 1
x <- rnorm(100, mu, sigma)
ggplot(data.frame(x=x), aes(x=x)) + geom_histogram(binwidth=0.5, color='grey')
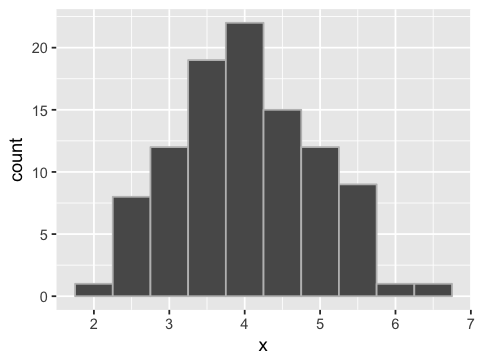
Likelihood
We have 100 samples independently drawn from the same normal
distribution - that is, the samples are IID (independent identically
distributed). The conditional probability of of a single sample
\(x_i\) is \(N(\mu, \sigma \mid x_i)\). Because the samples are
independent, the likelihood of seeing all 100 samples is just the
product of 100 such conditional probabilities.
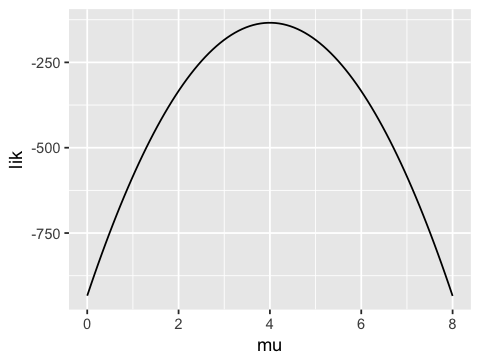
If we fix the standard deviation to be 1 and vary the mean, we see that
the likelihood function has a maximum near the true population mean
(\(\mu = 4\)). Hence we can get a good estimate of \(\mu\) by
maximizing the likelihood. Typically, a numerical optimization algorithm
is used to find the maximum - the value of \(\mu\) (or other
parameter) where this maximum occurs is the maximum likelihood
estimate (MLE) of the population parameter.
mus <- seq(0, 8, length.out = 100)
lik <- vector('numeric', length=100)
for (i in 1:100) {
lik[i] <- prod(dnorm(x, mean=mus[i], sd=1))
}
ggplot(data.frame(mu=mus, lik=lik), aes(x=mu, y=lik)) + geom_line()
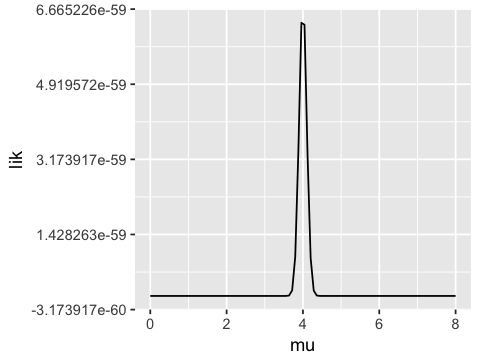
Working in log space
In practice, multiplying lots of small numbers to get a likelihood
results in underflow because of the discreteness of computer arithmetic.
So we work with log probabilities and sum them rather than take
products, but the basic idea of maximizing the (log) likelihood is the
same.
mus <- seq(0, 8, length.out = 100)
loglik <- vector('numeric', length=100)
for (i in 1:100) {
loglik[i] <- sum(log(dnorm(x, mean=mus[i], sd=1)))
}
ggplot(data.frame(mu=mus, lik=loglik), aes(x=mu, y=lik)) + geom_line()
Data with binary outcomes
We will try to predict admission to graduate school based on GRE, GPA
and class rank. Rank is the prestige of the undergraduate institution,
with 1 = most prestigious and 4 = least prestigious.
admit <- read.csv("http://stats.idre.ucla.edu/stat/data/binary.csv")
admit$rank <- as.factor(admit$rank)
'data.frame': 400 obs. of 4 variables:
$ admit: int 0 1 1 1 0 1 1 0 1 0 ...
$ gre : int 380 660 800 640 520 760 560 400 540 700 ...
$ gpa : num 3.61 3.67 4 3.19 2.93 3 2.98 3.08 3.39 3.92 ...
$ rank : Factor w/ 4 levels "1","2","3","4": 3 3 1 4 4 2 1 2 3 2 ...
| admit | gre | gpa | rank |
|---|
| 0 | 380 | 3.61 | 3 |
| 1 | 660 | 3.67 | 3 |
| 1 | 800 | 4.00 | 1 |
| 1 | 640 | 3.19 | 4 |
| 0 | 520 | 2.93 | 4 |
| 1 | 760 | 3.00 | 2 |
m <- glm(admit ~ gre + gpa + rank, data=admit, family="binomial")
Call:
glm(formula = admit ~ gre + gpa + rank, family = "binomial",
data = admit)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.6268 -0.8662 -0.6388 1.1490 2.0790
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -3.989979 1.139951 -3.500 0.000465 *
gre 0.002264 0.001094 2.070 0.038465 *
gpa 0.804038 0.331819 2.423 0.015388 *
rank2 -0.675443 0.316490 -2.134 0.032829 *
rank3 -1.340204 0.345306 -3.881 0.000104 *
rank4 -1.551464 0.417832 -3.713 0.000205 *
---
Signif. codes: 0 ‘*’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 499.98 on 399 degrees of freedom
Residual deviance: 458.52 on 394 degrees of freedom
AIC: 470.52
Number of Fisher Scoring iterations: 4
Waiting for profiling to be done...
| 2.5 % | 97.5 % |
|---|
| (Intercept) | -6.2716 | -1.7925 |
|---|
| gre | 0.0001 | 0.0044 |
|---|
| gpa | 0.1603 | 1.4641 |
|---|
| rank2 | -1.3009 | -0.0567 |
|---|
| rank3 | -2.0277 | -0.6704 |
|---|
| rank4 | -2.4000 | -0.7535 |
|---|
Test significance of GRE
wald.test(b = coef(m), Sigma = vcov(m), Terms = 2)
Wald test:
----------
Chi-squared test:
X2 = 4.3, df = 1, P(> X2) = 0.038
Test significance of GPA
wald.test(b = coef(m), Sigma = vcov(m), Terms = 3)
Wald test:
----------
Chi-squared test:
X2 = 5.9, df = 1, P(> X2) = 0.015
Test significance of Rank
wald.test(b = coef(m), Sigma = vcov(m), Terms = 4:6)
Wald test:
----------
Chi-squared test:
X2 = 20.9, df = 3, P(> X2) = 0.00011
Getting odds ratios
round(exp(cbind(OR = coef(m), confint(m))), 4)
Waiting for profiling to be done...
| OR | 2.5 % | 97.5 % |
|---|
| (Intercept) | 0.0185 | 0.0019 | 0.1665 |
|---|
| gre | 1.0023 | 1.0001 | 1.0044 |
|---|
| gpa | 2.2345 | 1.1739 | 4.3238 |
|---|
| rank2 | 0.5089 | 0.2723 | 0.9448 |
|---|
| rank3 | 0.2618 | 0.1316 | 0.5115 |
|---|
| rank4 | 0.2119 | 0.0907 | 0.4707 |
|---|
For every unit increase in GPA, the odds of being admitted increase by a
factor of 2.2345.
Making predictions
| admit | gre | gpa | rank |
|---|
| 0 | 380 | 3.61 | 3 |
| 1 | 660 | 3.67 | 3 |
| 1 | 800 | 4.00 | 1 |
| 1 | 640 | 3.19 | 4 |
| 0 | 520 | 2.93 | 4 |
| 1 | 760 | 3.00 | 2 |
new.data <- with(admit, data.frame(gre=mean(gre), gpa=mean(gpa), rank=factor(1:4)))
| gre | gpa | rank |
|---|
| 587.7 | 3.3899 | 1 |
| 587.7 | 3.3899 | 2 |
| 587.7 | 3.3899 | 3 |
| 587.7 | 3.3899 | 4 |
new.data$p <- predict(m, new.data, type="response")
new.data
| gre | gpa | rank | p |
|---|
| 587.7 | 3.3899 | 1 | 0.5166016 |
| 587.7 | 3.3899 | 2 | 0.3522846 |
| 587.7 | 3.3899 | 3 | 0.2186120 |
| 587.7 | 3.3899 | 4 | 0.1846684 |
Exercise
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked |
|---|
| 1 | 0 | 3 | Braund, Mr. Owen Harris | male | 22 | 1 | 0 | A/5 21171 | 7.2500 | | S |
| 2 | 1 | 1 | Cumings, Mrs. John Bradley (Florence Briggs Thayer) | female | 38 | 1 | 0 | PC 17599 | 71.2833 | C85 | C |
| 3 | 1 | 3 | Heikkinen, Miss. Laina | female | 26 | 0 | 0 | STON/O2. 3101282 | 7.9250 | | S |
| 4 | 1 | 1 | Futrelle, Mrs. Jacques Heath (Lily May Peel) | female | 35 | 1 | 0 | 113803 | 53.1000 | C123 | S |
| 5 | 0 | 3 | Allen, Mr. William Henry | male | 35 | 0 | 0 | 373450 | 8.0500 | | S |
| 6 | 0 | 3 | Moran, Mr. James | male | NA | 0 | 0 | 330877 | 8.4583 | | Q |
'data.frame': 891 obs. of 12 variables:
$ PassengerId: int 1 2 3 4 5 6 7 8 9 10 ...
$ Survived : int 0 1 1 1 0 0 0 0 1 1 ...
$ Pclass : int 3 1 3 1 3 3 1 3 3 2 ...
$ Name : chr "Braund, Mr. Owen Harris" "Cumings, Mrs. John Bradley (Florence Briggs Thayer)" "Heikkinen, Miss. Laina" "Futrelle, Mrs. Jacques Heath (Lily May Peel)" ...
$ Sex : chr "male" "female" "female" "female" ...
$ Age : num 22 38 26 35 35 NA 54 2 27 14 ...
$ SibSp : int 1 1 0 1 0 0 0 3 0 1 ...
$ Parch : int 0 0 0 0 0 0 0 1 2 0 ...
$ Ticket : chr "A/5 21171" "PC 17599" "STON/O2. 3101282" "113803" ...
$ Fare : num 7.25 71.28 7.92 53.1 8.05 ...
$ Cabin : chr "" "C85" "" "C123" ...
$ Embarked : chr "S" "C" "S" "S" ...
1. Fit a logistic regression model of survival against sex, age, and
passenger class (Pclass). Remember to convert to factors as appropriate.
Evaluate and interpret the impact of sex, age and passenger class on
survival.




