Statistics review 7: Correlation and regression¶
R code accompanying paper
Key learning points¶
- Correlation quantifies the strength of the linear relationship between a pair of variables
- Regression expresses the relationship in the form of an equation
suppressPackageStartupMessages(library(tidyverse))
options(repr.plot.width=4, repr.plot.height=3)
Scatter diagram¶
age <- c(60, 76, 81, 89, 44, 58, 55, 74, 45, 67, 72,
91, 76, 39, 71, 56, 77, 37, 64, 84)
urea <- c(1.099, 1.723, 2.054, 2.262, 1.686, 1.988, 1.131, 1.917, 1.548, 1.386,
2.617, 2.701, 2.054, 1.526, 2.002, 1.526, 1.825, 1.435, 2.460, 1.932)
ae <- data.frame(subject=1:20, age=age, urea=urea)
head(ae)
| subject | age | urea |
|---|---|---|
| 1 | 60 | 1.099 |
| 2 | 76 | 1.723 |
| 3 | 81 | 2.054 |
| 4 | 89 | 2.262 |
| 5 | 44 | 1.686 |
| 6 | 58 | 1.988 |
ggplot(ae, aes(x=age, y=urea)) + geom_point()

Correlation¶
Function to calculate Pearson’s correlatin¶
pearson <- function(x, y) {
xbar <- mean(x)
ybar <- mean(y)
sum((x - xbar)*(y-ybar))/sqrt(sum((x-xbar)^2)*sum((y-ybar)^2))
}
round(pearson(x=ae$age, y=ae$urea), 2)
Using built-in function¶
round(cor(x=ae$age, y=ae$urea, method = 'pearson'), 2)
Hypothesis test of correlation¶
cor.test(x=ae$age, y=ae$urea)
Pearson's product-moment correlation
data: ae$age and ae$urea
t = 3.3538, df = 18, p-value = 0.003535
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
0.2447935 0.8338342
sample estimates:
cor
0.6201371
Confidence interval for the population correlation coefficient¶
This is also provided by the built in test function.
z <- c(0.5 * log((1+0.62)/(1 - 0.62)))
se <- 1/sqrt(20 -3)
z.ci <- c(z - 1.96*se, z + 1.96*se)
round(z.ci, 2)
- 0.25
- 1.2
ci <- c((exp(2*z.ci[1]) - 1)/(exp(2*z.ci[1]) + 1),
(exp(2*z.ci[2]) - 1)/(exp(2*z.ci[2]) + 1))
round(ci, 2)
- 0.24
- 0.83
Misuse of correlation¶
Correlations may arise from third variable¶
z <- 3*rnorm(10)
x <- z + rnorm(10)
y <- z + rnorm(10)
df <- data.frame(x=x, y=y)
ggplot(df, aes(x=x, y=y)) + geom_point() + geom_smooth(method=lm) +
annotate("text", x=-4, y=5, label = paste("r =", round(cor(df$x, df$y), 2)))

Correlations can be misleading if relationships are non-linear¶
head(anscombe)
| x1 | x2 | x3 | x4 | y1 | y2 | y3 | y4 |
|---|---|---|---|---|---|---|---|
| 10 | 10 | 10 | 8 | 8.04 | 9.14 | 7.46 | 6.58 |
| 8 | 8 | 8 | 8 | 6.95 | 8.14 | 6.77 | 5.76 |
| 13 | 13 | 13 | 8 | 7.58 | 8.74 | 12.74 | 7.71 |
| 9 | 9 | 9 | 8 | 8.81 | 8.77 | 7.11 | 8.84 |
| 11 | 11 | 11 | 8 | 8.33 | 9.26 | 7.81 | 8.47 |
| 14 | 14 | 14 | 8 | 9.96 | 8.10 | 8.84 | 7.04 |
g1 <- ggplot(anscombe, aes(x=x1, y=y1)) +
geom_point() + geom_smooth(method=lm) +
annotate("text", x=6, y=9,
label = paste("r =", round(cor(anscombe$x1, anscombe$y1), 2)))
g2 <- ggplot(anscombe, aes(x=x2, y=y2)) +
geom_point() + geom_smooth(method=lm) +
annotate("text", x=6, y=9,
label = paste("r =", round(cor(anscombe$x2, anscombe$y2), 2)))
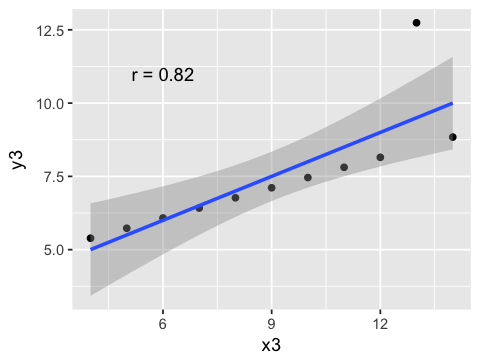
g3 <- ggplot(anscombe, aes(x=x3, y=y3)) +
geom_point() + geom_smooth(method=lm) +
annotate("text", x=6, y=11,
label = paste("r =", round(cor(anscombe$x3, anscombe$y3), 2)))
g4 <- ggplot(anscombe, aes(x=x4, y=y4)) +
geom_point() + geom_smooth(method=lm) +
annotate("text", x=10, y=11,
label = paste("r =", round(cor(anscombe$x4, anscombe$y4), 2)))
g1

g2

g3
g4
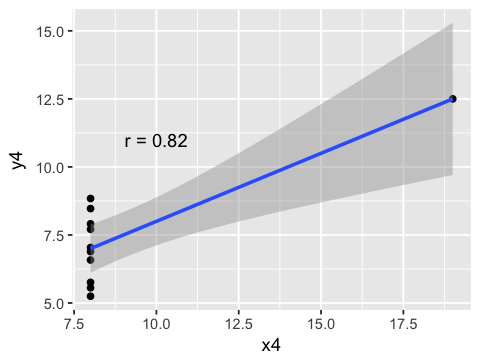
Correlations may arise from subgroups¶
m <- rnorm(10, 70, 4)
f <- rnorm(10, 65, 3.5)
x.m <- rnorm(10, 0)
x.f <- rnorm(10, 6)
df <- data.frame(x=c(x.f, x.m), height=c(f, m),
sex=c(rep("f", 10), rep("m", 10)))
ggplot(df, aes(x=x, y=height)) + geom_point() +
geom_smooth(method=lm) +
annotate("text", x=6, y=72,
label = paste("r =", round(cor(df$x, df$height), 2)))

Regression¶
x <- ae$age
xbar <- mean(x)
y <- ae$urea
ybar <- mean(y)
b <- sum((x - xbar)*(y - ybar))/sum((x-xbar)^2)
a <- ybar - b*xbar
round(c(a, b), 4)
- 0.7147
- 0.0172
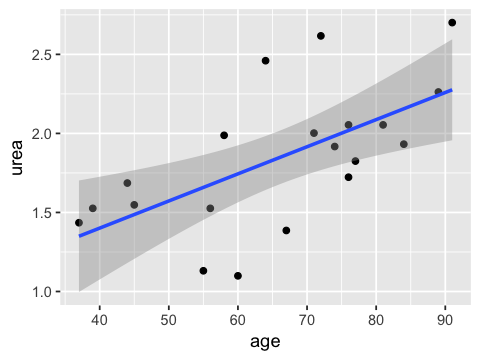
ggplot(ae, aes(x=age, y=urea)) + geom_point() +
geom_abline(aes(intercept=a, slope=b), color="blue")

Hypothesis tests and confidence intervals¶
fit <- lm(urea ~ age, data=ae)
summary(fit)
Call:
lm(formula = urea ~ age, data = ae)
Residuals:
Min 1Q Median 3Q Max
-0.64509 -0.21403 0.02789 0.16075 0.66703
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.714698 0.346129 2.065 0.05365 .
age 0.017157 0.005116 3.354 0.00354 **
---
Signif. codes: 0 ‘*’ 0.001 ‘’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.3606 on 18 degrees of freedom
Multiple R-squared: 0.3846, Adjusted R-squared: 0.3504
F-statistic: 11.25 on 1 and 18 DF, p-value: 0.003535
cor.test(ae$age, ae$urea)
Pearson's product-moment correlation
data: ae$age and ae$urea
t = 3.3538, df = 18, p-value = 0.003535
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
0.2447935 0.8338342
sample estimates:
cor
0.6201371
Analysis of variance¶

anova
fit.aov <- anova(fit)
fit.aov
| Df | Sum Sq | Mean Sq | F value | Pr(>F) | |
|---|---|---|---|---|---|
| age | 1 | 1.462673 | 1.4626726 | 11.24785 | 0.003535245 |
| Residuals | 18 | 2.340724 | 0.1300402 | NA | NA |
Residuals¶
ae$predicted <- predict(fit)
ae$residuals <- residuals(fit)
ggplot(ae, aes(x = age, y = urea)) +
geom_smooth(method = "lm", se = FALSE, color = "lightgrey") + # Plot regression slope
geom_segment(aes(xend = age, yend = predicted), alpha = .2) + # alpha to fade lines
geom_point() +
geom_point(aes(y = predicted), shape = 1)

Coefficient of determination¶
Regression_sum_of_squares <- fit.aov$'Sum Sq'[1]
Total_sum_of_squares <- fit.aov$'Sum Sq'[2]
round(c(Regression_sum_of_squares, Total_sum_of_squares), 4)
- 1.4627
- 2.3407
Explained variance¶
Age accounts for 38% of the total varia- tion in ln urea
r_squared = (Regression_sum_of_squares/Total_sum_of_squares)^2
round(r_squared, 2)
Prediction¶
newdata <- data.frame(age = seq(40, 90, 10))
newdata$urea.pred <- predict(fit, newdata)
newdata
| age | urea.pred |
|---|---|
| 40 | 1.400961 |
| 50 | 1.572526 |
| 60 | 1.744092 |
| 70 | 1.915658 |
| 80 | 2.087223 |
| 90 | 2.258789 |
ggplot(ae, aes(x=age, y=urea)) + geom_point() + geom_smooth(method=lm, )
names(fit)
- 'coefficients'
- 'residuals'
- 'effects'
- 'rank'
- 'fitted.values'
- 'assign'
- 'qr'
- 'df.residual'
- 'xlevels'
- 'call'
- 'terms'
- 'model'
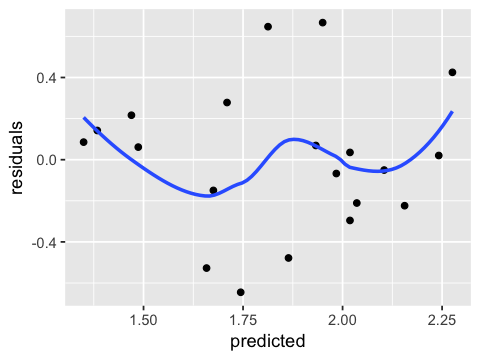
ggplot(ae, aes(x=predicted, y=residuals)) +
geom_point() + geom_smooth(se=FALSE)
geom_smooth() using method = 'loess'
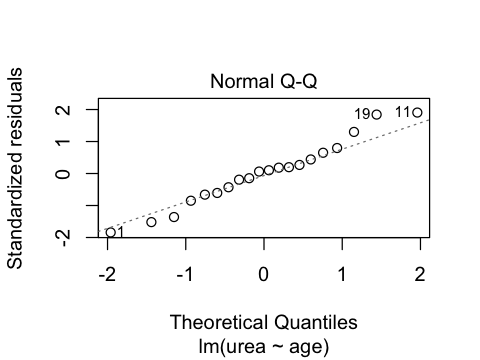
ggplot(ae, aes(sample=residuals)) + stat_qq()

Exercises¶
You may need to install the car package in R in the usual way:
install.packages("car")
suppressPackageStartupMessages(library(car))
head(Davis)
| sex | weight | height | repwt | repht |
|---|---|---|---|---|
| M | 77 | 182 | 77 | 180 |
| F | 58 | 161 | 51 | 159 |
| F | 53 | 161 | 54 | 158 |
| M | 68 | 177 | 70 | 175 |
| F | 59 | 157 | 59 | 155 |
| M | 76 | 170 | 76 | 165 |
1. Find the correlation between age and height for males and females
in the Davis data set.
2. Test if the correlation is significant.
3. Repeat exercises 1 and 2 taking for each sex spearately.
4. Show separate linear regressions of weight (y-axis) on height (x-axis) for each sex, either on the same plot (using different colors) or on different subplots.
5. What is the predicted weight of a female who is 165 cm tall?