Simulations and Statistical Inference¶
In [1]:
library(tidyverse)
── Attaching packages ─────────────────────────────────────── tidyverse 1.2.1 ──
✔ ggplot2 2.2.1 ✔ purrr 0.2.5
✔ tibble 1.4.2 ✔ dplyr 0.7.5
✔ tidyr 0.8.1 ✔ stringr 1.3.1
✔ readr 1.1.1 ✔ forcats 0.3.0
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
In [2]:
options(repr.plot.width=4, repr.plot.height=3)
Set random number seed for reproucibility¶
In [3]:
set.seed(42)
Generating numbers¶
Using the c function
In [4]:
c(1,2,3,5,8,13)
- 1
- 2
- 3
- 5
- 8
- 13
Using the : operator
In [5]:
1:10
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
In [6]:
10:1
- 10
- 9
- 8
- 7
- 6
- 5
- 4
- 3
- 2
- 1
Using the seq function
In [7]:
seq(1, 10)
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
In [8]:
seq(1, 10, 2)
- 1
- 3
- 5
- 7
- 9
In [9]:
seq(10, 1, -1)
- 10
- 9
- 8
- 7
- 6
- 5
- 4
- 3
- 2
- 1
In [10]:
seq(0, 1, length.out = 11)
- 0
- 0.1
- 0.2
- 0.3
- 0.4
- 0.5
- 0.6
- 0.7
- 0.8
- 0.9
- 1
Using rep¶
rep is useful for generating repeating data patterns.
In [11]:
rep(3, 5)
- 3
- 3
- 3
- 3
- 3
In [12]:
rep(1:3, times=5)
- 1
- 2
- 3
- 1
- 2
- 3
- 1
- 2
- 3
- 1
- 2
- 3
- 1
- 2
- 3
In [13]:
rep(1:3, each=5)
- 1
- 1
- 1
- 1
- 1
- 2
- 2
- 2
- 2
- 2
- 3
- 3
- 3
- 3
- 3
In [14]:
rep(1:3, length.out=10)
- 1
- 2
- 3
- 1
- 2
- 3
- 1
- 2
- 3
- 1
Using sample¶
Simulate n tooses of a coin
In [15]:
n <- 10
In [16]:
t1 <- sample(c('H', 'T'), n, replace = TRUE)
t1
- 'T'
- 'T'
- 'H'
- 'T'
- 'T'
- 'T'
- 'T'
- 'H'
- 'T'
- 'T'
In [17]:
table(t1)
t1
H T
2 8
Simulate n tosses of a biased coin
In [18]:
t2 <- sample(c('H', 'T'), n, replace = TRUE, prob = c(0.3, 0.7))
t2
- 'T'
- 'H'
- 'H'
- 'T'
- 'T'
- 'H'
- 'H'
- 'T'
- 'T'
- 'T'
In [19]:
table(t2)
t2
H T
4 6
Simulate n rolls of a 6-sided die
In [20]:
n <- 100
In [21]:
d1 <- sample(1:6, n, replace=TRUE)
d1
- 6
- 1
- 6
- 6
- 1
- 4
- 3
- 6
- 3
- 6
- 5
- 5
- 3
- 5
- 1
- 5
- 1
- 2
- 6
- 4
- 3
- 3
- 1
- 6
- 3
- 6
- 6
- 4
- 6
- 4
- 3
- 3
- 3
- 5
- 1
- 5
- 5
- 2
- 2
- 4
- 5
- 6
- 5
- 4
- 6
- 2
- 2
- 5
- 5
- 2
- 1
- 1
- 2
- 3
- 2
- 5
- 1
- 3
- 4
- 1
- 4
- 1
- 3
- 4
- 5
- 4
- 2
- 1
- 1
- 2
- 5
- 1
- 2
- 6
- 6
- 5
- 2
- 4
- 5
- 4
- 4
- 2
- 2
- 3
- 6
- 6
- 5
- 5
- 4
- 1
- 4
- 6
- 5
- 3
- 4
- 4
- 1
- 3
- 4
- 5
In [22]:
table(d1)
d1
1 2 3 4 5 6
16 14 15 18 20 17
Sampling without replacement. For example, if we wanted to assiggn 16 samples to treatment A or B at random such that exactly half had each treatment.
In [23]:
sample(rep(c('A', 'B'), each=8))
- 'A'
- 'A'
- 'B'
- 'A'
- 'B'
- 'A'
- 'B'
- 'B'
- 'A'
- 'B'
- 'B'
- 'A'
- 'A'
- 'B'
- 'B'
- 'A'
Random number generators¶
Disscrete distributionns¶
Sampling from a Bernoullli distribution returns TRUE for success and FALSE for failure.
In [24]:
rbernoulli(n=10, p=0.5)
- TRUE
- FALSE
- TRUE
- TRUE
- FALSE
- FALSE
- FALSE
- TRUE
- TRUE
- TRUE
In [25]:
as.integer(rbernoulli(n=10, p=0.5))
- 0
- 1
- 0
- 0
- 1
- 0
- 1
- 0
- 1
- 1
Sampling from a Binomial distribution returns the number of
successes in size trials for n experiments.
In [26]:
rbinom(n=10, p=0.5, size=5)
- 2
- 0
- 1
- 3
- 4
- 3
- 3
- 2
- 3
- 1
Sampling from a negative binomial distribution returns the number of
failures until size succcesses are observed for n
experiemnts.
In [27]:
rnbinom(n=10, size=5, prob=0.5)
- 2
- 5
- 6
- 7
- 6
- 6
- 5
- 1
- 0
- 9
Sampling from a Poisson distribution returns the number of successes
in n experiments if the average success rate per experiment is
lambda.
In [28]:
rpois(n=10, lambda = 3)
- 3
- 5
- 3
- 1
- 3
- 7
- 3
- 2
- 2
- 3
Note: We can give different parameters for each experiment in these distributios.
In [54]:
rpois(n=10, lambda=1:10)
- 2
- 2
- 3
- 5
- 6
- 3
- 6
- 8
- 9
- 9
Continuous distributions¶
Sampling from a stnadard uniform distribution.
In [29]:
runif(5)
- 0.649875837611035
- 0.336419132305309
- 0.0609497462864965
- 0.451310850214213
- 0.838755033444613
Sampling form a uniform distribuiotn,
In [30]:
runif(5, 90, 100)
- 95.7463733432814
- 93.5335037740879
- 95.4742607823573
- 98.9271859382279
- 94.8999057058245
Sampling from a stnadard normal distribution.
In [31]:
rnorm(5)
- -0.94773529068477
- 1.76797780755595
- 0.917327777548805
- -0.894775408854386
- -0.688165944958397
Looping¶
In [90]:
for (i in 1:10) {
print(mean(rnorm(10)))
}
[1] 0.2660734
[1] -0.09742615
[1] -0.3068994
[1] 0.2910631
[1] 0.1062726
[1] 0.05955272
[1] 0.4080255
[1] -0.1243275
[1] -0.779981
[1] -0.187303
Saving variabels generated in a loop
In [91]:
n <- 10
vars <- numeric(n)
for (i in 1:n) {
vars[i] <- mean(rnorm(10))
}
vars
- -0.25116755575127
- 0.708916638976887
- 0.461551515258198
- -0.475557761331935
- -0.0598841272831985
- 0.272569234838853
- 0.0543736652184016
- 0.135002662203851
- -0.190719191248592
- 0.0387989282162199
Using replicate¶
replicate is like rep but works for a function (such as a random
number geenerator)
In [32]:
replicate(3, rnorm(5))
| -0.9984839 | 0.15461594 | -0.88368060 |
| -2.1072414 | -0.80160087 | 0.07714041 |
| 0.8574190 | -0.04518119 | 0.83720136 |
| 1.1260757 | -1.03824075 | 0.09992556 |
| -0.1983684 | 1.55953311 | 0.30272834 |
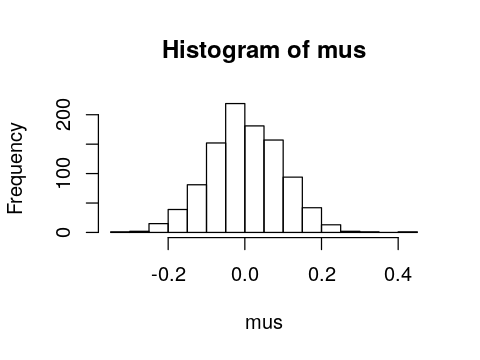
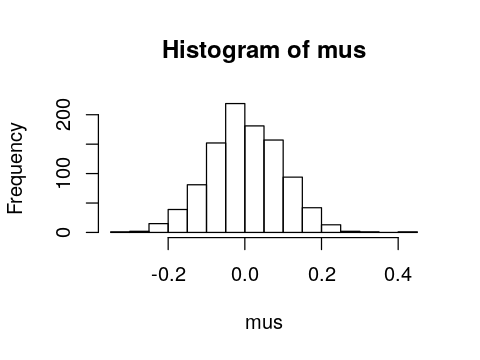
replicate is quite useful for simulations. For exampe, suppose we
want to know the distribution of the sample mean if we sampled 100
numbers from the standard normal distribution 1,000 times.
In [ ]:
n_expts <- 1000
n <- 100
Using for loop¶
In [86]:
set.seed(123)
mus <- numeric(n_expts)
for (i in 1:n_expts) {
mus[i] <- mean(rnorm(n))
}
hist(mus)
Makign a data.frame of simulated data¶
Let’s simulate the following experiment.
- There are 10 subjects in Group A and 10 subjects in Group B with ranodm PIDs from 10000-99999
- We measure 5 genes in each subject. The genes have the same
distribution for each subject, but different genes have differnt
distribtutions:
- gene1 \(\sim N(10, 1)\)
- gene2 \(\sim N(11, 2)\)
- gene3 \(\sim N(12, 3)\)
- gene4 \(\sim N(13, 4)\)
- gene5 \(\sim (N(14, 5)\)
In [48]:
replicate(5, rnorm(3, 1:5, 1))
| 0.6536426 | 1.334889 | 1.5432302 | 0.6573478 | 2.249920 |
| 1.3585636 | 2.068446 | 0.8936747 | 4.2271172 | 3.146313 |
| 3.4023661 | 2.388056 | 2.6394386 | 1.5861398 | 2.943069 |
In [75]:
n <- 10
n_genes <- 5
min_pid <- 10000
max_pid <- 99999
groupings <- c('A', 'B')
n_groups <- length(groupings)
gene_mus <- 10:14
gene_sigmas <- 1:5
pad_width <- 3
In [76]:
pids <- sample(min_pid:max_pid, n_groups*n)
groups <- sample(rep(groupings, each=n))
genes <- t(replicate(n_groups*n, rnorm(n_genes, gene_mus, gene_sigmas)))
gene_names <- paste('gene', str_pad(1:n_genes, width = pad_width, pad='0'), sep='')
colnames(genes) <- gene_names
In [77]:
df <- data.frame(
pid = pids,
grp = groups,
genes
)
In [58]:
sample_n(df, 3)
| pid | grp | gene001 | gene002 | gene003 | gene004 | gene005 | |
|---|---|---|---|---|---|---|---|
| 16 | 38729 | B | 1.530708 | 8.613943 | 10.617613 | 9.215513 | 9.880912 |
| 13 | 22215 | B | 14.545609 | 10.625483 | 6.288087 | 9.741466 | 9.624057 |
| 14 | 89658 | A | 13.008224 | 14.321570 | 8.115676 | 12.608564 | 7.962775 |
Breakdown of simjulation¶
Set up simulation confiugration parameters.
In [70]:
n <- 10
n_genes <- 5
min_pid <- 10000
max_pid <- 99999
groupings <- c('A', 'B')
n_groups <- length(groupings)
gene_mus <- 10:14
gene_sigmas <- 1:5
pad_width <- 3
Create unique PIDs for each subject
In [62]:
pids <- sample(min_pid:max_pid, n_groups*n)
Assign a group to each subject at ranodm
In [67]:
groups <- sample(rep(groupings, each=n))
Make up 5 genes from different distributions for each subject
In [68]:
genes <- t(replicate(n_groups*n, rnorm(n_genes, gene_mus, gene_sigmas)))
Make nice names for each gene
In [71]:
gene_names <- paste('gene', str_pad(1:n_genes, width = pad_width, pad='0'), sep='')
Assign names to gene columns
In [72]:
colnames(genes) <- gene_names
Create data.frame to storre simulated data
In [73]:
df <- data.frame(
pid = pids,
grp = groups,
genes
)
Peek into data.frame
In [74]:
sample_n(df, 3)
| pid | grp | gene001 | gene002 | gene003 | gene004 | gene005 | |
|---|---|---|---|---|---|---|---|
| 16 | 72254 | B | 10.726324 | 9.749522 | 9.772832 | 19.65021 | 13.226052 |
| 18 | 21661 | B | 10.448878 | 12.689833 | 13.292832 | 19.66553 | 14.330836 |
| 14 | 91408 | B | 9.125949 | 11.009273 | 13.052141 | 14.95065 | 8.745456 |