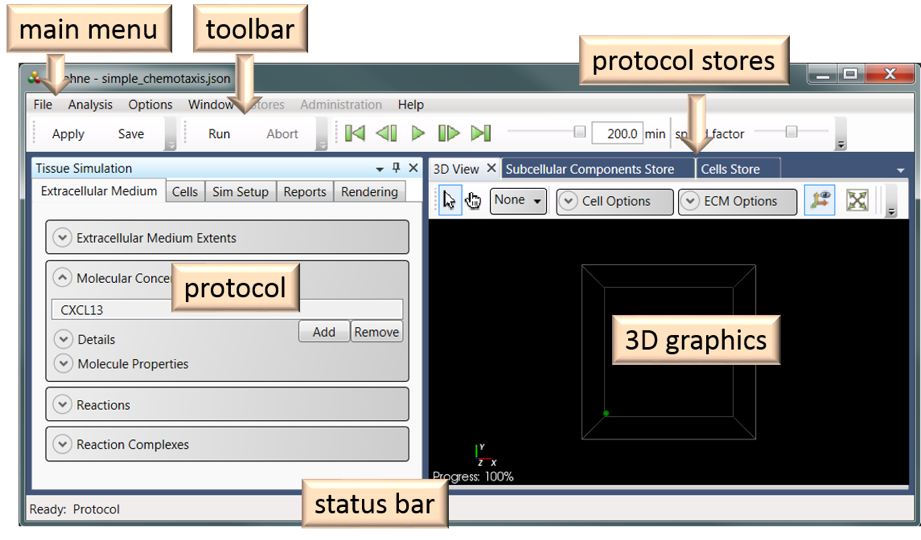
The Graphical User Interface (GUI)
The Graphical User Interface (GUI) is comprised of six functional units (Figure 13): Main Menu, Toolbar, Protocol, 3D Graphics, Protocol Stores, and Status Bar. The simulation protocol, 3D graphics, and protocol stores windows can be undocked (and re-docked).
Protocol
The Protocol section of the GUI is where the user specifies the protocol for the simulation experiment.
Cells
Populations
The Populations section of the Cells tab (Figure 3) allows the user to specify the types, numbers, and initial positions of cells in the simulation experiment.
Cells are added as populations of the same cell type and the user selects the initial placement of the population of cells in the ECM. Placement is either random, using uniform or normal distributions (Figure 4), or explicit, with the user typing or pasting cell coordinates into a table.
Properties
The Properties section displays the properties of the cell type of the selected cell population. The cell properties are subdivided into subsections for Movement, Chemistry, and Transitions, as outlined in the Cell biology section earlier (Figure 5). The Properties section allows the user to make local changes to the cell properties (e.g., changing a reaction rate constant or the chemotactic transduction parameter). These local changes only persist in the current protocol and it’s simulation experiments and do not affect other protocols that use the same cell type. However, the Save Cell to Cells Library button can be used if the user wishes to make the local changes persist beyond the current protocol, as was discussed in the Hierarchies of Persistence section earlier. The Movement submenu is shown in (Figure 5a). The Chemistry, and Transitions submenus are as shown in Figure 6-Figure 10.
The parameters of some cell properties can be distributed, rather than being constant (the same) for all cells in the population. An example of this is illustrated in Figure 5b, where a Weibull distribution has been selected for the transduction constant. In this case, instead of having all cells in the population (same cell type) have the same (Constant) chemotactic transduction constant, the value for each cell is randomly selected from the specified distribution. In addition to the transduction constant, the drag coefficient and stochastic motion force parameter can also be distributed.
Discrete distributions (Categorical or Poisson) can be selected for the initial cell states (differentiation, division, and death). This is illustrated in Figure 11, where a categorical distribution has been selected for the initial death state of the cells in the population. In this case, the categorical distribution will be used to randomly assign the initial death states of cells as alive or dead.
Removal. The Removal section allows the user to specify whether dead cells will be removed from the simulation immediately or stochastically (Figure 12). The latter choice can be helpful for comparison to experimental data. With the default settings (Constant distribution, Value = 0), dead cells will be removed immediately. A non-zero constant value specifies that dead cells will be removed after the specified number of minutes. If a distribution (Gamma, Negative Exponential, Uniform, or Weibull) is chosen, the time interval (minutes) before removal after cell death is a number randomly chosen from the specified distribution.
Interaction Parameters. This section contains system parameters for cell-cell interactions. The casual user is discouraged from modifying these parameters
Sim Setup
The parameters in the Sim Setup tab allows the user to specify the experiment name and description, time configuration parameters, and boundary condition for the simulation. Examples of the latter two are shown in Figure 13. All time units are specified in units of minutes. The total time is the length of the simulation. The rendering interval determines how often the 3D graphics are refreshed and data are written to the HDF5 database. The sampling interval determines how often data are written to the report files. The integration step is the fixed time step used in an Euler integration scheme. It is recommended that users do not change this parameter. The repetitions parameter specifies how many replicate simulations should be run. This can be important when the simulation has stochastic components, such as with transitions. The global random seed is fixed for each protocol, unless modified by the user. This parameter enables reproducibility of stochastic simulations. If the same (stochastic) simulation is run multiple times with the same global random seed, then the results will be identical.
There are two choices possible for the Boundary Condition option: Zero Flux and Toroidal. The Zero Flux option means that there is no flux of ECM molecules through the sides of the simulation space – all molecules remain in the simulation space. Similarly, there is a repulsive force at the simulation boundaries that tends to keep cells in the simulation space. This force can be likened to the affect of cells surrounding the simulation space that would tend to oppose motion of the cell out of the simulation space.
The Toroidal boundary condition means that cells leaving from the left face re-enter from the right face of the simulation space and cells leaving through the top face re-enter through the bottom face and, similarly, for the front and back faces of the simulation space. Molecular diffusion will occur as if the right and left faces are the same and, similarly, for the right/left and top/bottom face.
Reports
The Reports feature in Daphne provides the capability to record data to file during the simulation for post-simulation analysis. With this feature, users can specify which data should be recorded and with what frequency the data should be recorded. Figure 14 provides a summary of the organization of the Reports menu.
Reporting details are specified in the sub-menus shown in Figure 14 and as detailed below.
Report Settings: The user can specify the root name for data files. Depending on the selections made, the reported data will be organized in one or more files.
Molecules in Extracellular Medium: For each molecule type in the extracellular medium, the user can specify reporting of the mean concentration (‘mean’) and/or reporting on the spatially-distributed concentration (‘full’), which records the concentration at each grid point. The user can also select to report on the gradient of the molecular concentration at each grid point with the ‘full + gradient’ setting.
Cell Information: For each cell population, the user can specify reporting of cell positions, velocities, and forces.
Cell State: For each cell population, the user can specify reporting on the death, division, and differentiation states of the cells.
Molecules by cell: For each cell population and molecular population in the plasma membrane or cytosol, the user can specify reporting of the mean concentration or the mean concentration and the gradient through the ‘Molecules in membrane’ and ‘Molecules in cytosol’ sub-menus. In the ‘ECM probe at cell location’, the user can specify reporting extracellular molecular concentrations in the vicinity of the cell. This feature is useful for probing the local environment of the cell.
The frequency of reporting is specified in the ‘Sim Setup’ menu with the ‘sampling interval’ choice.
Rendering
Rendering refers to the visual representation of cells or molecular populations in the ECM. Our objective was to allow users to design a detailed rendering scheme independently of the Daphne code, which can then be distributed as a file and used in many different simulations. In other words, the look and feel of the visualization can be customized by the user without the need to work with the code base.
With the cell rendering capabilities provide in Daphne, the user can color cells by state (differentiation, division, or death), generation (number of divisions), cell type, or cell population. The user can toggle between render options during the simulation (while in the Pause mode) or during playback of the simulation, while preserving the data associated with each option. In other words, a previously-set-up option is restorable. A rendering scheme can be edited and saved as a file. It can be loaded into a rendering dictionary in the simulation while any protocol is open. Entities and colors are matched via labels in the dictionary. A system-wide default color is used for entities if no color entry exists.
The graphical user interface provides a choice for the rendering option (e.g. rendering by type, state, generation etc.) and allows the user to customize the various color selections (Figure 15).
3D view
The 3D View window (Figure 16a) gives a representation of the cell locations and states with perspective. Various options are available in the ribbon menu above the 3D View window (Figure 17).
Various mouse controls allow the user to rotate, pan, or scale the view when the Navigation Tool is selected (Figure 17a). Holding the left mouse button down and dragging the cursor horizontally along the 2D x-axis rotates the view around the 2D y-axis. Similarly, holding the left mouse button down and dragging the cursor along the direction of the 2D y-axis will rotate the view about the 2D x-axis. Holding the right mouse button down while dragging the cursor up (down) causes the view to zoom in (zoom out). Panning is achieved by dragging the cursor while holding down the SHIFT and left mouse buttons down.
The cell selector and mode option (Figure 17b) will be discussed in greater detail in section White Hand tool
As discussed in the Rendering section above, cells can be colored according to their type, population, or state. The Cell Options tab above the 3D window offers an easy way to change the cell rendering choices on the fly (Figure 17c). The user can also select shape of the rendered cell with the Cell Shape option. In addition to the default Spheres choice, the user can choose Polygon or Point (Figure 16b-d). The advantage of the latter two is that they are computationally less expensive than rendering as a sphere and can greatly reduce the simulation time, especially when there are many cells in the simulation. As with the other rendering options, the user can make changes during the simulation (while in the Pause mode) or during playback of the simulation.
The Orientation axes in the corner of the 3D View window can be toggled on or off with the Orientation axis visibility control (Figure 17d).
The Reset camera option (Figure 17e) camera can be reset to the default orientation with the 3D x-axis horizontal, the y-axis vertical, and the z-axis out of the screen.
The user can select a different background color with the Choose background option (Figure 17f).
The 3D view can be exported to various file formats (JPEG, Bitmap, PNG, and TIFF) with the Export option (Figure 17g).
Protocol Stores
The Subcellular Components Store is a repository of all the subcellular components in, or available to be added into, the protocol (Figure 18). The Cells Store is a repository of all cells in, or available to be added into, the protocol (Figure 19). More items can be made available to these stores through the Stores option in the Main Menu.
The Subcellular Components and Cells Stores are one level up from the Protocol level in the Hierarchy of Persistence (see section Hierarchies of Persistence). Entities, such as cells or molecules, can be modified here, independently of the same entities in the Protocol level or one level up in the User Store
Main Menu
The File submenu in the Main Menu (Figure 1) contains options for loading and saving protocols, importing and exporting SBML files, loading and exporting past experiments, and exiting.
The Analysis submenu contains options for plotting cell population dynamics and cell pedigrees.
The Options submenu allows the user to manage preferences and data storage options.
The Window submenu can be used to open windows, such as the Protocol or 3D View, if the user has closed them.
The Stores submenu allows the user to get or send items, such as molecules and cells, among the various Stores (Daphne, User, and Protocol). An example of the Molecules Store is shown in Figure 20.
The Administration submenu allows the user to view all the items in the User store. When Daphne is first installed, the User store is a copy of the published Daphne store. Users cannot modify items in the Daphne store, but they can edit their User store through the Administration submenu.
Status Bar
The Status bar indicates the status of the system. The Ready: Protocol status indicates the system is available for edits to the protocol and the Running and Paused statuses indicate when the simulation is running or paused.
Simulation playback
Users have the capability to replay simulation experiments in Daphne. Data are saved to an HDF5 database file during the simulation and these experiment files can be reloaded for playback. The user can control the playback speed and export the 3D visualization to an AVI file. As is the case during the simulation, the user can pause the playback and use the White Hand tool to probe cells for quantitative information about their internal dynamics. All the options for cell rendering (e.g., cell coloring based on differentiation state) are also available during playback. The playback capability is provided in addition to the customized reporting of simulation data to text files that is available to users.