Software Design¶
Daphne was designed to be a user-friendly desktop application for immunologists and vaccine researchers who wish to simulate multi-scale germinal center dynamics for insight into host-pathogen interactions or optimal vaccine design. Given these requirements, Daphne was developed for 64-bit .NET systems, taking advantage of the .NET class libraries to manage complexity and to develop familiar user interfaces for modern desktop applications.
The system is organized as three loosely coupled layers, consisting of the graphical user interface (GUI), the simulation engine, and the persistence/data model. The GUI was developed using Windows Presentation Foundation (WPF) as a base, and using the third party libraries Actipro for enhanced controls, the Visualization Toolkit (VTK) for 3D rendering, and SciChart for charting data. The simulation engine was developed largely in C#, with focused sections developed in unmanaged C++ for efficiency and access to high-performance numerical libraries. Communication between the GUI and the simulation engine follows the Model/View/View-Controller (MVVC) framework commonly used in .NET applications. The persistence/data model layer uses a combination of Hierarchical Data Format 5 (HDF5) for efficient storage of complex array-based data, and JavaScript Object Notation (JSON) for readable and portable configuration files.
Daphne instantiates a simulation based on protocol files. The protocol files describe all configuration entities in a simulated experiment: cell populations, cells, extra-cellular space (ECS), molecules, reactions, reaction complexes and their respective properties. A loaded simulation mirrors the protocol’s configuration entities by creating matching simulation entities with the properties described in the protocol. Running a simulation will generate two main types of output files: report files that serve for post-run analysis, and an HDF5 file that allows playing back the simulation as an animated movie.
Design Principles¶
The maturation of the germinal center over time is complex at multiple levels, from cells to processes to the individual molecular populations that underlie each cellular process. Consequently, setting up a computational experiment to simulate some biological scenario requires the flexibility to configure a hierarchy of dependencies – for example, inclusion of a population of B cells requires information about how their cell division processes are to be implemented, which might in turn depend on information about the population of molecules that regulate the cell cycle. Since specific aspects of this information may need to be changed depending on the biological scenario being simulated, the code has to be flexible enough to cope with run-time wiring of object dependencies. A related aspect is the need, at run-time, to be able to replace or “swap” specific biological modules with a variant. To meet this need, we evolved the C# code base to increase modularity and flexibility, via an increased level of decoupling and abstraction. Even with swappable modules, model selection and calibration can be time-consuming in individual (or agent)-based models, due to the computationally intensive nature of such simulations. It therefore makes sense in specific contexts, to develop reduced models that capture key aspects of the planned simulation for testing and parameter range finding, and use this to inform the development of the full model. Finally, we describe an approach to model parameter estimation, and how all these will be integrated into a calibration workbench in MSI.
In order to increase flexibility for modeling biological complexity needed both at compile-time and at run-time, the computational framework has been revised for loose coupling. Loose coupling is widely understood to be desirable for software design because it improves maintainability and extensibility, enables late binding (run-time rather than compile-time resolution of object methods), makes testing easier and allows code development in parallel by more than one programmer. However, designing for loose coupling and composability demands a high degree of abstraction, and is a departure from the pattern of creating concrete classes “on demand” to meet a need that developers often turn to.
One aspect of a loosely coupled design is the use of abstract and orthogonal sets of classes. At some level, all biological dynamics can be expressed as changes in either physical forces or chemical reactions. While it would be impractical to describe immunology at this level, these concepts form the foundation of the simulation framework we also describe how familiar immunological concepts can be built on top of these abstractions.
The implementation of the base classes (underlined) for handling chemical reactions follows the following scheme. Reactions are dynamic interactions of MolecularPopulations containing ScalarFields, occurring in Compartments whose topology is defined by a Manifold. The Manifold and ScalarField classes play particularly critical roles, since they map directly to the mathematical concepts of manifold and scalar field. Scalar fields describe concentration and flux information for molecular populations, and their associated manifolds determine the spatial distribution of these values. This mapping enforces a rigorous implementation for the operations of addition, multiplication, scalar multiplication, gradient and Laplacian, allowing the simulation to carry out the necessary operations for chemical reaction-diffusion systems.
Currently there are three inter-convertible implementations of the Manifold/ScalarField mapping, a moment expansion (ME) formalism, a more explicit lattice interpolation (IL) scheme, and a point manifold (PM) implementation. The first two provide a useful trade-off between memory space (ME) and accuracy (IL) as needed in different biological compartments. The third implementation (PM) is convenient for well-mixed compartments, such as in the Vat Reaction Complex workbench, where the dynamics have no spatial dependence.
Compartments may nest other compartments; the nested compartments are treated as the boundary of the parent compartment. By treating the germinal center tissue environment, individual cell plasma membranes and cytosols as separate nested compartments, and specifying how each compartment handles bulk reaction-diffusion processes as well as bulk-boundary reaction-diffusion processes, all reactions in the simulation are handled using a common execution path. Within each compartment, processes are just named collections of reactions, and so cellular processes such as cell division are also handled using the same scheme. In addition, some processes also incorporate the generation or response to physical forces, for example, cell locomotion, where a force influencing the cell’s velocity is generated by the chemotaxis reactions together with physical contact with other cells or environmental structures.
The abstraction provided by the class design described enables composability. Each of these base classes may have derived classes, but the derived classes use the same interface specification, ensuring that any class of a particular type may be swapped with another of the same type without affecting the ability of the code to compile and run. As described, the class design relies on object composition by aggregation – each class may have a “wiring diagram” or object graph showing how it is built up from simpler classes, which may also be compositional. Concrete instantiation at multiple of these dependencies leads to tight coupling and less flexibility in the ability of the end-user to configure immunological scenarios in detail. Instead, these dependencies are included by “injection” into object constructors, allowing us to delay specification of each object to a single point in the code, the object composition root. Configuration files specified by the end-user at run-time can in turn inform the specification of dependencies that is needed in the object composition root.
The use of abstract base classes allows swapping of instances within the same family. However, to integrate the code across multiple scales, we also want to be able to swap instances of classes from different families. For example, we may want to test a module such as a cell division process sometimes in isolation to evaluate the reaction model itself (e.g. does it generate the appropriate oscillatory behavior with the correct periodicity?), sometimes in the context of a cell to evaluate how it interacts with other cellular processes, and sometimes within a population of cells to collect statistics for a simulation of cell birth and death events in some environmental context. This is implemented by the use of interfaces, in this particular case with an IDynamic interface that guarantees that all classes with this interface will implement a void step(double dt) method in order that we can run simulation entities at multiple scales (e.g. process, cell, populations of cells) independently. By making processes, cells and the simulation extend the IDynamic interface, a common framework for testing system dynamics can be developed, regardless of whether the system is a single process, a cell or an entire simulation with a tissue context and potentially many distinct cell populations.
Hierarchies of Persistence¶
Daphne comes with a full set of calibrated biological entities (molecules, genes, reactions, reaction complexes, and cells) that can be deployed in simulation experiments (Protocols). These entities can be used as-is, modified by the user, or the user may choose to create new entities of their own design. In particular, users may want to modify the parameters of predefined entities in a single experiment or a related set of experiments – for example, users may want to make adjustments to entity predefined parameter values to run simulation experiments to test hypotheses. This flexibility in parameter values requires similar flexibility in the graphical user interface and a well-defined persistence scheme.
The persistence scheme is comprised of 4 levels of information storage:
- Daphne store – special role, updated by the publisher
- User store – updated by the user
- Protocol store – updated by the user
- Entity – updated by the user
We can think of the levels as a typical hierarchy of parent and child levels. Daphne is the parent of User, User is a child of Daphne, and so on. The items numbered below detail changes to information stores at these levels and communication of changes between levels.
- Local edits are made at Entity store (e.g. reaction rate constant) via the GUI
- Users can choose to push changes up to the Protocol store
- Changes only communicate down to other Entity stores under the Protocol on REQUEST.
- Users can choose to push changes up to User store.
- Changes only communicate down to other Protocol stores under User on REQUEST.
- Push changes up to Daphne store – a reserved function that requires super-user privileges. Daphne store changes get pushed to Users on first use and also on REQUEST thereafter.
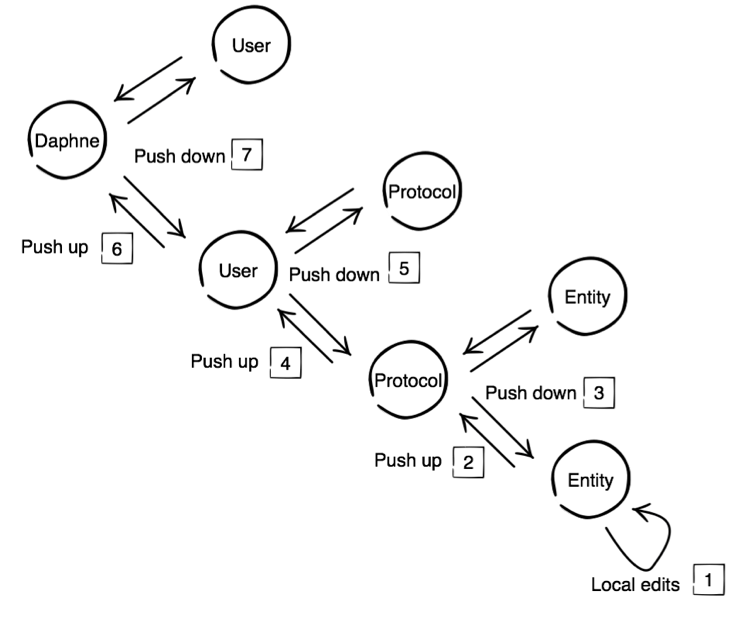
Figure 1: Schematic for persistence and communication of changes. Note that Protocol + Entity are stored in the same protocol (JSON-formatted) file. User and Daphne will be stored in their own JSON-formatted
Implementation¶
All editing and creating of new entities happens at the Entity level. Changes can get pushed between neighboring levels. The upper levels, Protocol, User, Daphne, are ‘stores’ that hold available entities. The stores can get updated by pushing entities in the applicable direction. The Daphne level plays a special role in that regular users can push data from it to the User level but not vice versa.
The Daphne level can be considered a super-user level. It gets updated by its publisher only. The User level is a copy or subset of Daphne, copied upon first system start or by request. Daphne and User contain a single instance of data. However, there can and will be more than one protocol in the Protocol level. Each protocol stores the entities that play a role in the protocol. The Entity level stores the actual instances of entities with their parameter values as they are used in the experiment.
The entities are the same as what are in the component library: genes, molecules, reactions, reaction complexes, cells, etc., and also the building blocks of each, e.g. molecular populations, behaviors (death, division), and so on.
We allow the user to select an entity and change it locally only in the Entity level, then push it back to Protocol and from there to the User level upon demand. To allow for local edits in the Entity level all entities that need to be editable have to store their values rather than hold a reference to another location. In addition they are identifiable through a unique identifier / reference, e.g. a GUID. The identifier / reference serves for the purpose of pushing into and out of a level. This was a departure from the previous implementation that used references throughout.
Protocol contains entities selected from User, or it can have its own custom entities that were created through the GUI in the Entity level and pushed back into Protocol. Similarly, Entity can contain verbatim or edited versions of entities that came from Protocol or new, custom-made ones. Unless the user pushes data back from a child to its parent level the two can contain different entities or the same entities with different values.
A protocol is self-contained. It can be instantiated into a simulation without any knowledge of what is in User or Daphne.
A change / edit in Entity stays local, i.e. only the underlying entity has its value data updated. Upon request, one can push Entity changes back to Protocol. We also have the ability to push changes from Protocol to Entity. The system will display a notification when a newer, updated version is available with the option to update all entity instances in Entity or none. Similarly, one can push changes from Protocol to User. This should not automatically affect other protocols.
Synopsis¶
- pushes can occur between levels in both directions except User – Daphne
- pushes from a higher to a lower level, e.g. User to Protocol, affect all instances in the lower level or one selected instance
- initialization of User from Daphne
- value data (instead of references) in Entity level
- edits only in Entity level
- notifications of newer data
- protocols are self-contained
- levels saved are as individual files
This persistence scheme provides the flexibility for users to customize their own biological entities and carry out hypothesis testing.
Workbenches¶
A full simulation of the Germinal Center requires simulation of the complex dynamics of biological entities, such as B cells, T cells, and FDC, interacting with each other and with the environment. Dynamics in Daphne are built on low-level processes: diffusion reaction systems, stochastic transitions, and Newtonian forces. The diffusion reaction systems occur both in the environment of the extracellular medium and in the internal environment of the cell. Calibration of a full germinal center system is therefore a difficult task, because the cells are, themselves, complex systems.
In order to facilitate calibration of the Germinal Center system, the Daphne software has been designed to be able to support Workbench environments. These Workbenches provide the capability for users to explore the behavior of smaller subsystems and calibrate them prior to their use in the full simulation. These environments can expose smaller systems, such as sets of reactions, single cells, and cell-cell interactions, and provide the capability for the user to rapidly explore behavior as a function of the adjustable parameters.
Although the user could implement these smaller systems in the full simulation environment, the workbenches can provide simpler environments that facilitate exploration of the parameter space. This can achieved through specialized displays that respond to interactive adjustment of parameters. Once calibrated, these systems can be pushed to the User store for use in a full simulation.