Unsupervised and Supervised Learning¶
In [2]:
library(tidyverse)
Loading tidyverse: ggplot2
Loading tidyverse: tibble
Loading tidyverse: tidyr
Loading tidyverse: readr
Loading tidyverse: purrr
Loading tidyverse: dplyr
Conflicts with tidy packages ---------------------------------------------------
filter(): dplyr, stats
lag(): dplyr, stats
In [24]:
library(pheatmap)
In [3]:
options(repr.plot.width=6, repr.plot.height=6)
Toy data set¶
In [4]:
head(iris)
| Sepal.Length | Sepal.Width | Petal.Length | Petal.Width | Species |
|---|---|---|---|---|
| 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 5.0 | 3.6 | 1.4 | 0.2 | setosa |
| 5.4 | 3.9 | 1.7 | 0.4 | setosa |
Note: If you want graphics based on ggplot2, install the
`GGally <https://cran.r-project.org/web/packages/GGally/index.html>`__
package and use the ggpairs function. This has not been installed on
the OIT Docker containers, so we use the simpler pairs function from
base graphics.
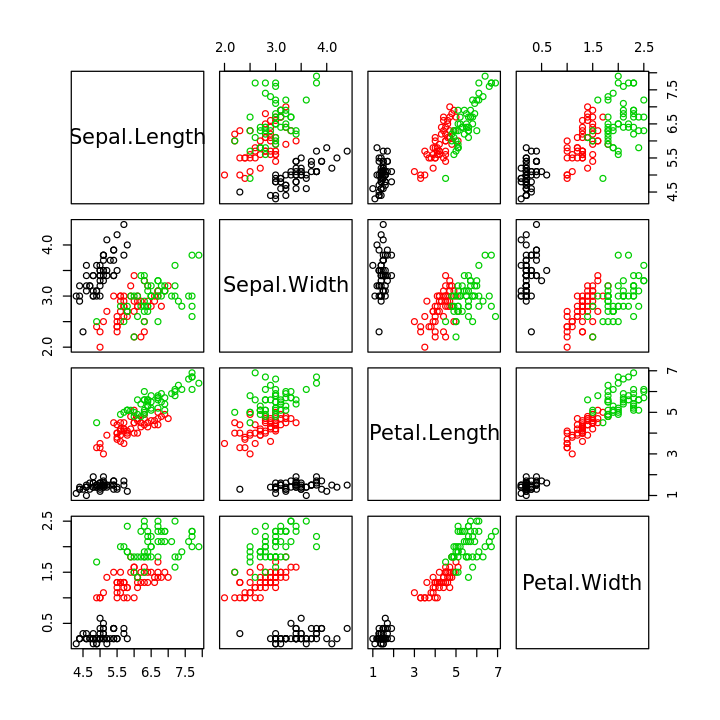
library(GGally)
ggpairs(iris, aes(colour = Species))
In [5]:
pairs(iris[,1:4], col=iris$Species)
Unsupervised Learning¶
Ordination (Dimension reduction)¶
PCA¶
In [6]:
pca <- as.data.frame(prcomp(iris[,1:4], center=TRUE, scale=TRUE, rank=2)$x)
In [7]:
dim(pca)
- 150
- 2
In [8]:
pca <- bind_cols(iris, pca)
In [9]:
head(pca)
| Sepal.Length | Sepal.Width | Petal.Length | Petal.Width | Species | PC1 | PC2 |
|---|---|---|---|---|---|---|
| 5.1 | 3.5 | 1.4 | 0.2 | setosa | -2.257141 | -0.4784238 |
| 4.9 | 3.0 | 1.4 | 0.2 | setosa | -2.074013 | 0.6718827 |
| 4.7 | 3.2 | 1.3 | 0.2 | setosa | -2.356335 | 0.3407664 |
| 4.6 | 3.1 | 1.5 | 0.2 | setosa | -2.291707 | 0.5953999 |
| 5.0 | 3.6 | 1.4 | 0.2 | setosa | -2.381863 | -0.6446757 |
| 5.4 | 3.9 | 1.7 | 0.4 | setosa | -2.068701 | -1.4842053 |
In [10]:
options(repr.plot.width=5.5, repr.plot.height=4)
In [11]:
ggplot(pca, aes(x=PC1, y=PC2, color=Species)) +
geom_point()
Data type cannot be displayed:

MDS¶
In [12]:
dist(iris[1:5,1:4])
1 2 3 4
2 0.5385165
3 0.5099020 0.3000000
4 0.6480741 0.3316625 0.2449490
5 0.1414214 0.6082763 0.5099020 0.6480741
In [13]:
mds <- as.data.frame(cmdscale(dist(iris[,1:4]), k = 2))
In [14]:
dim(mds)
- 150
- 2
In [15]:
mds <- bind_cols(mds, iris)
In [16]:
head(mds)
| V1 | V2 | Sepal.Length | Sepal.Width | Petal.Length | Petal.Width | Species |
|---|---|---|---|---|---|---|
| -2.684126 | 0.3193972 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| -2.714142 | -0.1770012 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| -2.888991 | -0.1449494 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| -2.745343 | -0.3182990 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| -2.728717 | 0.3267545 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
| -2.280860 | 0.7413304 | 5.4 | 3.9 | 1.7 | 0.4 | setosa |
In [17]:
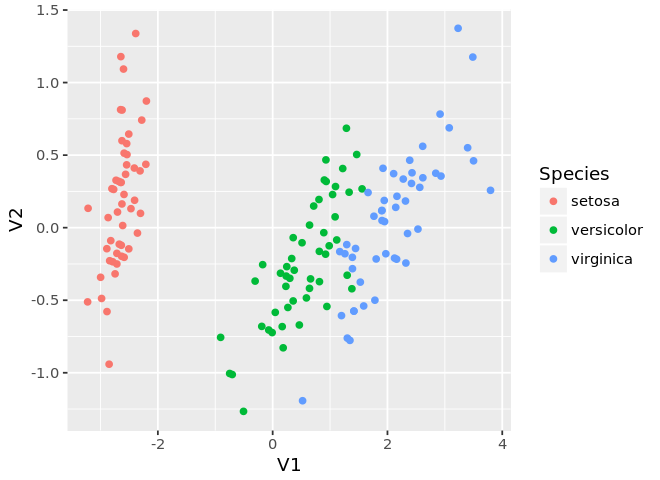
ggplot(mds, aes(x=V1, y=V2, color=Species)) +
geom_point()
Data type cannot be displayed:
Clustering¶
Note: If you want graphics based on ggplot2, install the
`gdendro <https://cran.r-project.org/web/packages/ggdendro/vignettes/ggdendro.html>`__
package and use the ggdendrogram function. This has not been
installed on the OIT Docker containers, so we use the simpler plot
function from base graphics.
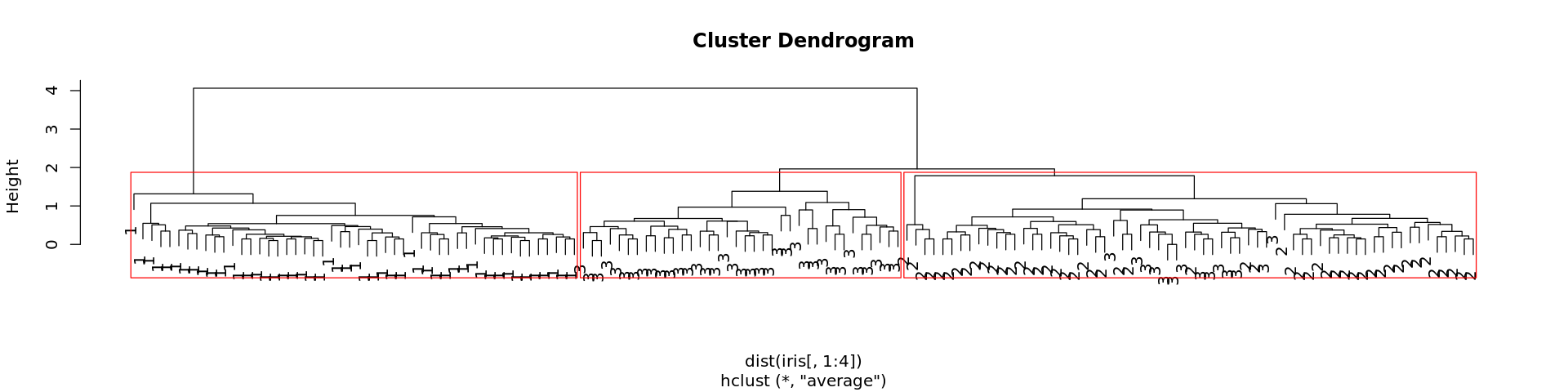
hc <- hclust(dist(iris[,1:4]), 'complete')
ggdendrogram(hc, rotate = FALSE, size = 2)
Agglomertive clustering¶
In [18]:
options(repr.plot.width=16, repr.plot.height=4)
In [19]:
tree <- hclust(dist(iris[,1:4]), method='average')
In [20]:
tree$labels <- as.integer(iris$Species)
In [21]:
plot(tree)

In [22]:
plot(tree)
rect.hclust(tree, k=3, border = "red")
Dendrograms are also constructed as part of visualizing a heatmapm¶
In [33]:
?pheatmap
In [29]:
options(repr.plot.width=5, repr.plot.height=6)
In [32]:
pheatmap(iris[,1:4], show_rownames = FALSE)

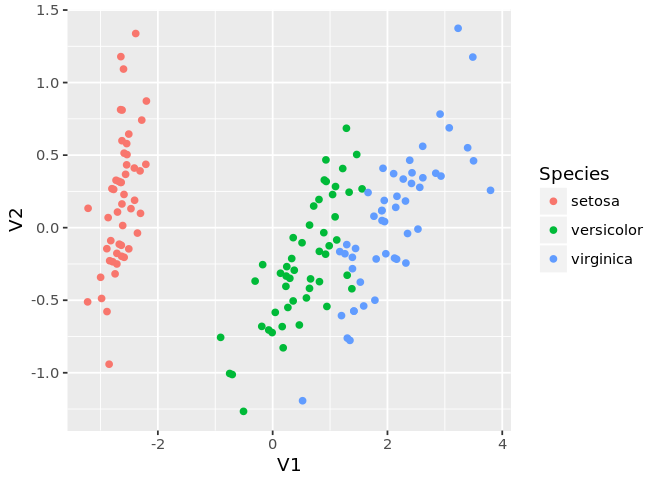
Visualize clusters on MDS plot¶
In [23]:
ggplot(mds, aes(x=V1, y=V2, color=Species)) +
geom_point() +
guides(color=guide_legend(title='Species'))
Data type cannot be displayed:
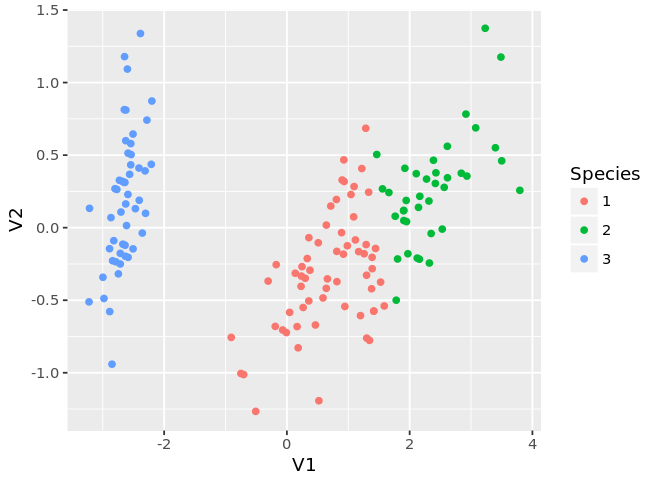
In [24]:
z <- cutree(tree, k = 3)
In [25]:
ggplot(mds, aes(x=V1, y=V2, color=as.factor(z))) +
geom_point() +
guides(color=guide_legend(title='Species'))
Data type cannot be displayed:

K-means clustering¶
In [26]:
km <- kmeans(iris[,1:4], centers=3)
In [27]:
km
K-means clustering with 3 clusters of sizes 62, 38, 50
Cluster means:
Sepal.Length Sepal.Width Petal.Length Petal.Width
1 5.901613 2.748387 4.393548 1.433871
2 6.850000 3.073684 5.742105 2.071053
3 5.006000 3.428000 1.462000 0.246000
Clustering vector:
[1] 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3
[38] 3 3 3 3 3 3 3 3 3 3 3 3 3 1 1 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
[75] 1 1 1 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 1 2 2 2 2 1 2 2 2 2
[112] 2 2 1 1 2 2 2 2 1 2 1 2 1 2 2 1 1 2 2 2 2 2 1 2 2 2 2 1 2 2 2 1 2 2 2 1 2
[149] 2 1
Within cluster sum of squares by cluster:
[1] 39.82097 23.87947 15.15100
(between_SS / total_SS = 88.4 %)
Available components:
[1] "cluster" "centers" "totss" "withinss" "tot.withinss"
[6] "betweenss" "size" "iter" "ifault"
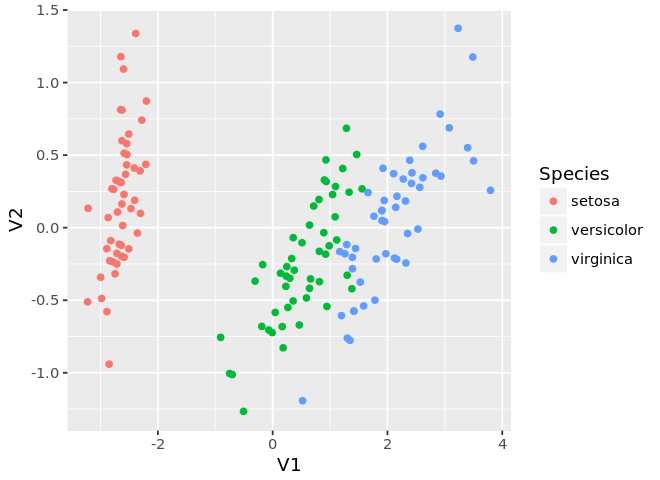
Visualize clusters on MDS plot¶
In [28]:
ggplot(mds, aes(x=V1, y=V2, color=Species)) +
geom_point() +
guides(color=guide_legend(title='Species'))
Data type cannot be displayed:
In [29]:
ggplot(mds, aes(x=V1, y=V2, color=as.factor(km$cluster))) +
geom_point() +
guides(color=guide_legend(title='Species'))
Data type cannot be displayed:
Supervised Learning¶
If you need to tune the model parameters, you will need to construct an additional validation data set.
In [30]:
idx <- sample(1:nrow(iris), 50)
In [31]:
idx
- 113
- 57
- 22
- 141
- 94
- 140
- 44
- 103
- 62
- 29
- 16
- 41
- 87
- 8
- 53
- 146
- 21
- 46
- 3
- 18
- 150
- 15
- 25
- 60
- 109
- 38
- 34
- 130
- 81
- 144
- 26
- 19
- 80
- 96
- 40
- 121
- 92
- 119
- 27
- 11
- 75
- 149
- 114
- 20
- 129
- 73
- 137
- 14
- 47
- 51
In [32]:
iris.test <- iris[idx,]
iris.train <- iris[-idx,]
In [33]:
library(nnet)
In [34]:
fit <- multinom(Species ~ ., data=iris.train)
# weights: 18 (10 variable)
initial value 109.861229
iter 10 value 9.488101
iter 20 value 5.718048
iter 30 value 5.607010
iter 40 value 5.566651
iter 50 value 5.564816
iter 60 value 5.559930
final value 5.559325
converged
In [35]:
fit
Call:
multinom(formula = Species ~ ., data = iris.train)
Coefficients:
(Intercept) Sepal.Length Sepal.Width Petal.Length Petal.Width
versicolor 15.30068 -4.714006 -8.950189 12.66693 2.132277
virginica -19.12182 -6.891088 -15.500148 20.84077 18.004415
Residual Deviance: 11.11865
AIC: 31.11865
In [36]:
pred <- predict(fit, newdata = iris.test)
In [37]:
mds.test <- mds[idx,]
In [38]:
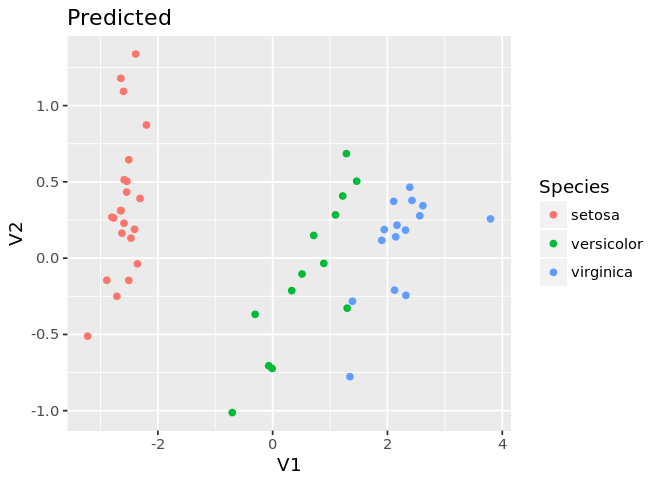
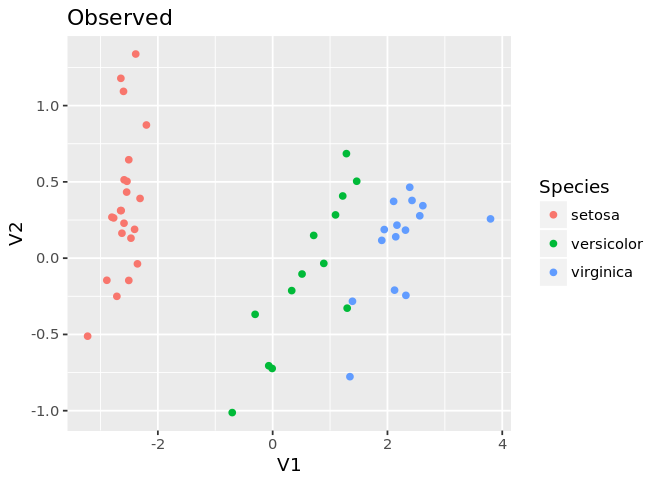
ggplot(mds.test, aes(x=V1, y=V2, color=as.factor(pred))) +
geom_point() +
guides(color=guide_legend(title='Species')) +
labs(title='Observed')
Data type cannot be displayed:
In [39]:
ggplot(mds.test, aes(x=V1, y=V2, color=as.factor(Species))) +
geom_point() +
guides(color=guide_legend(title='Species')) +
labs(title='Predicted')
Data type cannot be displayed: