Machine Learning with sklearn¶
This is mostly a tutorial to illustrate how to use scikit-learn to
perform common machine learning pipelines. It is NOT meant to show how
to do machine learning tasks well - you should take a machine learning
course for that.
%matplotlib inline
import itertools as it
import numpy as np
import pandas as pd
from pandas import DataFrame, Series
import matplotlib.pyplot as plt
Resources¶
Example¶
We will try to separate rocks from mines using this [data set](https://archive.ics.uci.edu/ml/datasets/Connectionist+Bench+(Sonar,+Mines+vs.+Rocks).
From the description provided:
Data Set Information:
The file "sonar.mines" contains 111 patterns obtained by bouncing sonar signals off a metal cylinder at various angles and under various conditions. The file "sonar.rocks" contains 97 patterns obtained from rocks under similar conditions. The transmitted sonar signal is a frequency-modulated chirp, rising in frequency. The data set contains signals obtained from a variety of different aspect angles, spanning 90 degrees for the cylinder and 180 degrees for the rock.
Each pattern is a set of 60 numbers in the range 0.0 to 1.0. Each number represents the energy within a particular frequency band, integrated over a certain period of time. The integration aperture for higher frequencies occur later in time, since these frequencies are transmitted later during the chirp.
The label associated with each record contains the letter "R" if the object is a rock and "M" if it is a mine (metal cylinder). The numbers in the labels are in increasing order of aspect angle, but they do not encode the angle directly.
df = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/undocumented/connectionist-bench/sonar/sonar.all-data', header=None, prefix='X')
df.shape
(208, 61)
The last column are labels - make it a category¶
df.rename(columns={'X60':'Label'}, inplace=True)
df.Label = df.Label.astype('category')
df.head()
| X0 | X1 | X2 | X3 | X4 | X5 | X6 | X7 | X8 | X9 | ... | X51 | X52 | X53 | X54 | X55 | X56 | X57 | X58 | X59 | Label | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.0200 | 0.0371 | 0.0428 | 0.0207 | 0.0954 | 0.0986 | 0.1539 | 0.1601 | 0.3109 | 0.2111 | ... | 0.0027 | 0.0065 | 0.0159 | 0.0072 | 0.0167 | 0.0180 | 0.0084 | 0.0090 | 0.0032 | R |
| 1 | 0.0453 | 0.0523 | 0.0843 | 0.0689 | 0.1183 | 0.2583 | 0.2156 | 0.3481 | 0.3337 | 0.2872 | ... | 0.0084 | 0.0089 | 0.0048 | 0.0094 | 0.0191 | 0.0140 | 0.0049 | 0.0052 | 0.0044 | R |
| 2 | 0.0262 | 0.0582 | 0.1099 | 0.1083 | 0.0974 | 0.2280 | 0.2431 | 0.3771 | 0.5598 | 0.6194 | ... | 0.0232 | 0.0166 | 0.0095 | 0.0180 | 0.0244 | 0.0316 | 0.0164 | 0.0095 | 0.0078 | R |
| 3 | 0.0100 | 0.0171 | 0.0623 | 0.0205 | 0.0205 | 0.0368 | 0.1098 | 0.1276 | 0.0598 | 0.1264 | ... | 0.0121 | 0.0036 | 0.0150 | 0.0085 | 0.0073 | 0.0050 | 0.0044 | 0.0040 | 0.0117 | R |
| 4 | 0.0762 | 0.0666 | 0.0481 | 0.0394 | 0.0590 | 0.0649 | 0.1209 | 0.2467 | 0.3564 | 0.4459 | ... | 0.0031 | 0.0054 | 0.0105 | 0.0110 | 0.0015 | 0.0072 | 0.0048 | 0.0107 | 0.0094 | R |
5 rows × 61 columns
Exploratory data analysis¶
We can use simple plots to get some idea of what the data looks like
- Is the separation between rocks and mines obvious?
- How correlated are the variables?
- Do we need to standardize?
- Are there outliers?
from pandas.tools.plotting import andrews_curves, parallel_coordinates
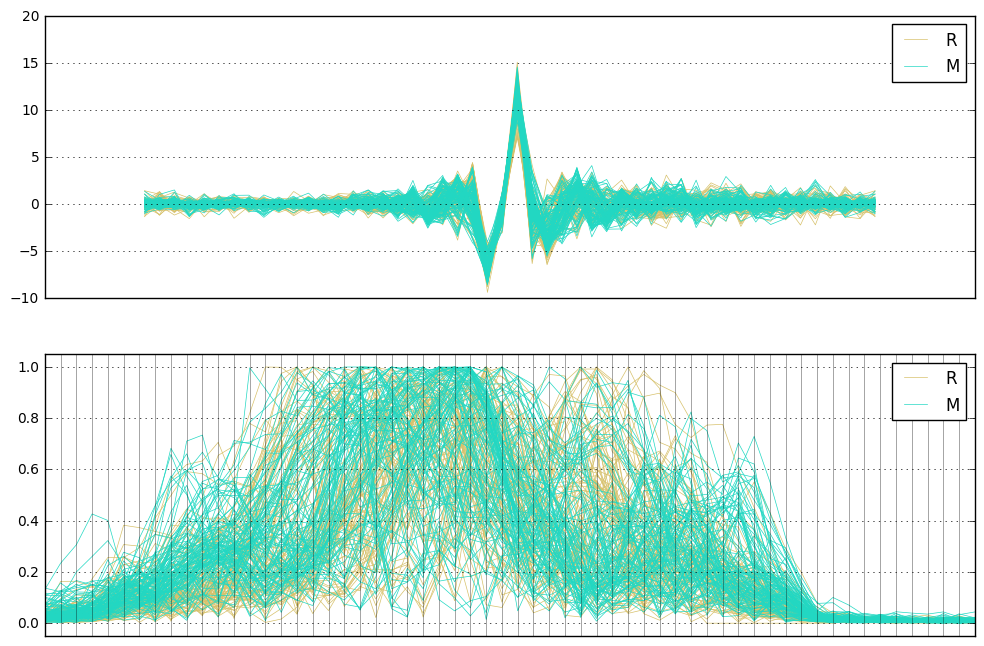
fig, axes = plt.subplots(2,1,figsize=(12,8))
andrews_curves(df, 'Label', samples=50, linewidth=0.5, ax=axes[0])
axes[0].set_xticks([])
parallel_coordinates(df, 'Label', linewidth=0.5, ax=axes[1],
axvlines_kwds={'linewidth': 0.5, 'color': 'black', 'alpha':0.5})
axes[1].set_xticks([])
axes[1].margins(0.05)
pass
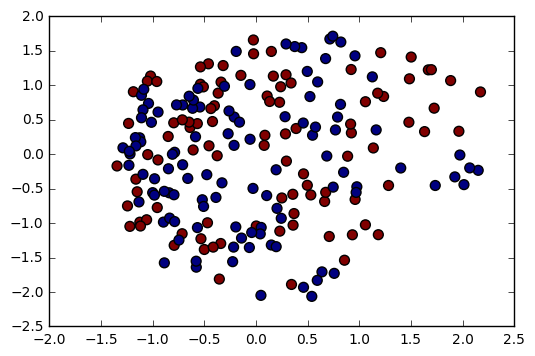
from sklearn.manifold import MDS
mds = MDS(n_components=2)
mds_data = mds.fit_transform(df.ix[:, :-1])
plt.scatter(mds_data[:, 0], mds_data[:, 1], c=df.Label.cat.codes, s=50);
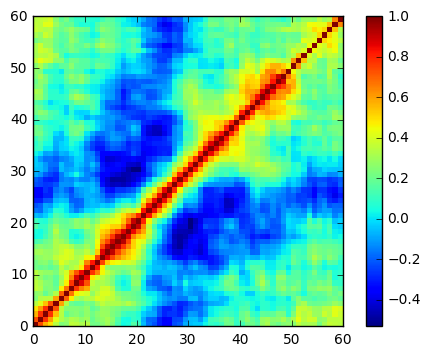
heatmap = plt.pcolor(df.corr(), cmap='jet')
plt.colorbar(heatmap)
plt.gca().set_aspect('equal')
df.plot.box(figsize=(12,4), xticks=[])
pass

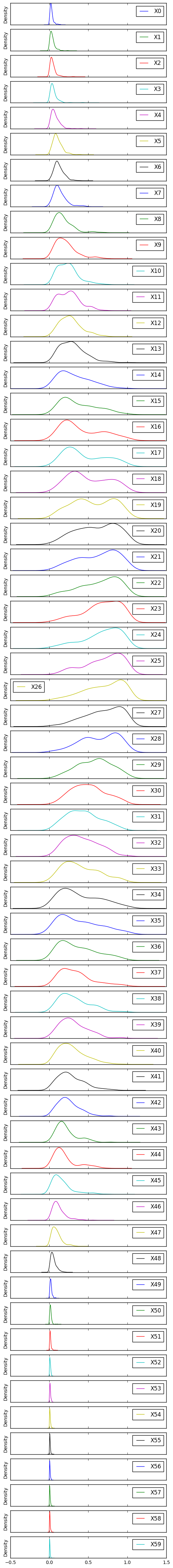
df.plot.density(figsize=(6, 60), subplots=True, yticks=[])
pass
Preprocessing¶
Box plots suggest we should standardize the data¶
from sklearn.preprocessing import StandardScaler, RobustScaler
data, labels = df.ix[:, :-1], df.ix[:, -1]
data.head(3)
| X0 | X1 | X2 | X3 | X4 | X5 | X6 | X7 | X8 | X9 | ... | X50 | X51 | X52 | X53 | X54 | X55 | X56 | X57 | X58 | X59 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.0200 | 0.0371 | 0.0428 | 0.0207 | 0.0954 | 0.0986 | 0.1539 | 0.1601 | 0.3109 | 0.2111 | ... | 0.0232 | 0.0027 | 0.0065 | 0.0159 | 0.0072 | 0.0167 | 0.0180 | 0.0084 | 0.0090 | 0.0032 |
| 1 | 0.0453 | 0.0523 | 0.0843 | 0.0689 | 0.1183 | 0.2583 | 0.2156 | 0.3481 | 0.3337 | 0.2872 | ... | 0.0125 | 0.0084 | 0.0089 | 0.0048 | 0.0094 | 0.0191 | 0.0140 | 0.0049 | 0.0052 | 0.0044 |
| 2 | 0.0262 | 0.0582 | 0.1099 | 0.1083 | 0.0974 | 0.2280 | 0.2431 | 0.3771 | 0.5598 | 0.6194 | ... | 0.0033 | 0.0232 | 0.0166 | 0.0095 | 0.0180 | 0.0244 | 0.0316 | 0.0164 | 0.0095 | 0.0078 |
3 rows × 60 columns
data_scaled = DataFrame(StandardScaler().fit_transform(data), columns=data.columns)
data_scaled.head(3)
| X0 | X1 | X2 | X3 | X4 | X5 | X6 | X7 | X8 | X9 | ... | X50 | X51 | X52 | X53 | X54 | X55 | X56 | X57 | X58 | X59 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | -0.399551 | -0.040648 | -0.026926 | -0.715105 | 0.364456 | -0.101253 | 0.521638 | 0.297843 | 1.125272 | 0.021186 | ... | 0.595283 | -1.115432 | -0.597604 | 0.680897 | -0.295646 | 1.481635 | 1.763784 | 0.069870 | 0.171678 | -0.658947 |
| 1 | 0.703538 | 0.421630 | 1.055618 | 0.323330 | 0.777676 | 2.607217 | 1.522625 | 2.510982 | 1.318325 | 0.588706 | ... | -0.297902 | -0.522349 | -0.256857 | -0.843151 | 0.015503 | 1.901046 | 1.070732 | -0.472406 | -0.444554 | -0.419852 |
| 2 | -0.129229 | 0.601067 | 1.723404 | 1.172176 | 0.400545 | 2.093337 | 1.968770 | 2.852370 | 3.232767 | 3.066105 | ... | -1.065875 | 1.017585 | 0.836373 | -0.197833 | 1.231812 | 2.827246 | 4.120162 | 1.309360 | 0.252761 | 0.257582 |
3 rows × 60 columns
If there are gross outliers, we can use a robust routine¶
data_robust = DataFrame(RobustScaler().fit_transform(data), columns=data.columns)
data_robust.head(3)
| X0 | X1 | X2 | X3 | X4 | X5 | X6 | X7 | X8 | X9 | ... | X50 | X51 | X52 | X53 | X54 | X55 | X56 | X57 | X58 | X59 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | -0.126126 | 0.200000 | 0.217949 | -0.581931 | 0.528726 | 0.096125 | 0.642271 | 0.538267 | 1.163123 | 0.182309 | ... | 0.750000 | -0.920635 | -0.310433 | 0.723288 | -0.037736 | 1.595142 | 1.791822 | 0.385185 | 0.390977 | -0.387097 |
| 1 | 1.013514 | 0.682540 | 1.282051 | 0.619315 | 0.896746 | 2.476155 | 1.486320 | 2.646482 | 1.330279 | 0.665714 | ... | -0.112903 | -0.317460 | -0.066158 | -0.493151 | 0.238994 | 1.983806 | 1.197026 | -0.133333 | -0.180451 | -0.165899 |
| 2 | 0.153153 | 0.869841 | 1.938462 | 1.601246 | 0.560868 | 2.024590 | 1.862517 | 2.971685 | 2.987903 | 2.775925 | ... | -0.854839 | 1.248677 | 0.717557 | 0.021918 | 1.320755 | 2.842105 | 3.814126 | 1.570370 | 0.466165 | 0.460829 |
3 rows × 60 columns
Dimension reduction¶
from sklearn.decomposition import PCA
data.shape
(208, 60)
pca = PCA()
data_scaled_pca = DataFrame(pca.fit_transform(data_scaled), columns=data.columns)
data_scaled.shape
(208, 60)
v = pca.explained_variance_ratio_
vc = v.cumsum()
DataFrame(list(zip(it.count(), v, vc)), columns=['pc', 'explained', 'cumsum']).head(10)
| pc | explained | cumsum | |
|---|---|---|---|
| 0 | 0 | 0.203466 | 0.203466 |
| 1 | 1 | 0.188972 | 0.392438 |
| 2 | 2 | 0.085500 | 0.477938 |
| 3 | 3 | 0.056792 | 0.534730 |
| 4 | 4 | 0.050071 | 0.584800 |
| 5 | 5 | 0.040650 | 0.625450 |
| 6 | 6 | 0.032790 | 0.658240 |
| 7 | 7 | 0.030465 | 0.688705 |
| 8 | 8 | 0.025660 | 0.714364 |
| 9 | 9 | 0.024911 | 0.739275 |
Let’s just use the principal components that explain at least 95% of total variance¶
n_comps = 1 + np.argmax(vc > 0.95)
n_comps
30
data_scaled_pca = data_scaled_pca.ix[:, :n_comps]
data_scaled_pca.shape
(208, 30)
data_scaled_pca.head()
| X0 | X1 | X2 | X3 | X4 | X5 | X6 | X7 | X8 | X9 | ... | X20 | X21 | X22 | X23 | X24 | X25 | X26 | X27 | X28 | X29 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.921168 | -1.370893 | -1.666476 | 0.837913 | -1.057324 | 1.712504 | 1.785716 | -1.581264 | 0.335418 | -1.028065 | ... | -1.208238 | 0.723202 | 0.304876 | 0.120470 | -0.458567 | -0.021847 | -1.089710 | 0.096606 | 0.168123 | -0.753434 |
| 1 | -0.480125 | 7.586388 | -1.275734 | 3.859346 | 2.121112 | -2.186818 | -1.742764 | 1.517061 | 0.307933 | -1.341882 | ... | -2.388110 | 0.021429 | -0.145524 | -0.246021 | 0.117770 | 0.704112 | -0.052387 | -0.240064 | -0.178744 | -0.554605 |
| 2 | 3.859228 | 6.439860 | -0.030635 | 5.454599 | 1.552060 | 1.181619 | -1.820138 | -1.495929 | -1.152459 | -1.006030 | ... | -1.740823 | -2.000942 | -0.295682 | 1.931963 | 0.758036 | -0.113901 | 0.964319 | 0.214707 | 0.527529 | -0.033003 |
| 3 | 4.597419 | -3.104089 | -1.785344 | -1.115908 | -2.785528 | -2.072673 | 2.084530 | 1.707289 | 0.452390 | -1.117318 | ... | -0.685825 | 1.307367 | -0.662918 | 1.142591 | -0.352601 | -0.491193 | -0.061186 | 0.150725 | 1.389191 | 0.642030 |
| 4 | -0.533868 | 1.849847 | -0.860097 | 3.302076 | 2.808954 | -0.783945 | 0.362657 | 0.812621 | 0.184578 | -0.023594 | ... | 0.503340 | 0.258970 | 0.253982 | 1.199262 | -0.165722 | -0.041342 | -0.589311 | -0.500720 | -1.549835 | -0.783667 |
5 rows × 30 columns
df_pca = pd.concat([data_scaled_pca, labels], axis=1)
df_pca.shape
(208, 31)
Classification¶
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = \
train_test_split(data_scaled_pca, labels, test_size=0.33, random_state=42)
from sklearn.linear_model import LogisticRegression
lr = LogisticRegression()
lr.fit(X_train, y_train)
LogisticRegression(C=1.0, class_weight=None, dual=False, fit_intercept=True,
intercept_scaling=1, max_iter=100, multi_class='ovr', n_jobs=1,
penalty='l2', random_state=None, solver='liblinear', tol=0.0001,
verbose=0, warm_start=False)
Accuracy score¶
lr.score(X_test, y_test)
0.78260869565217395
Using Support Vector Classifier and Grid Search¶
from sklearn.svm import SVC
from sklearn.model_selection import GridSearchCV
parameters = [{'kernel': ['rbf'], 'gamma': [1e-3, 1e-4],
'C': [1, 10, 100, 1000]},
{'kernel': ['linear'], 'C': [1, 10, 100, 1000]}]
Do grid search with parallel jobs¶
clf = GridSearchCV(SVC(C=1), parameters, cv=5, scoring='accuracy', n_jobs=-1)
clf.fit(X_train, y_train.codes)
pass
clf.best_params_
{'C': 1000, 'gamma': 0.001, 'kernel': 'rbf'}
clf.best_score_
0.76978417266187049
clf.score(X_test, y_test.codes)
0.88405797101449279
from sklearn.metrics import classification_report
y_true, y_pred = y_test.codes, clf.predict(X_test)
print(classification_report(y_true, y_pred))
precision recall f1-score support
0 0.88 0.92 0.90 38
1 0.90 0.84 0.87 31
avg / total 0.88 0.88 0.88 69
Using a Random Forests Classifier¶
from sklearn.ensemble import RandomForestClassifier
X_train, X_test, y_train, y_test = \
train_test_split(data_scaled, labels, test_size=0.33, random_state=42)
parameters = [{'n_estimators': list(range(25, 201, 25)),
'max_features': list(range(2, 15, 2))}]
clf = GridSearchCV(RandomForestClassifier(), parameters,
cv=5, scoring='accuracy', n_jobs=-1)
clf.fit(X_train, y_train.codes)
pass
clf.best_params_
{'max_features': 6, 'n_estimators': 150}
clf.score(X_test, y_test.codes)
0.85507246376811596
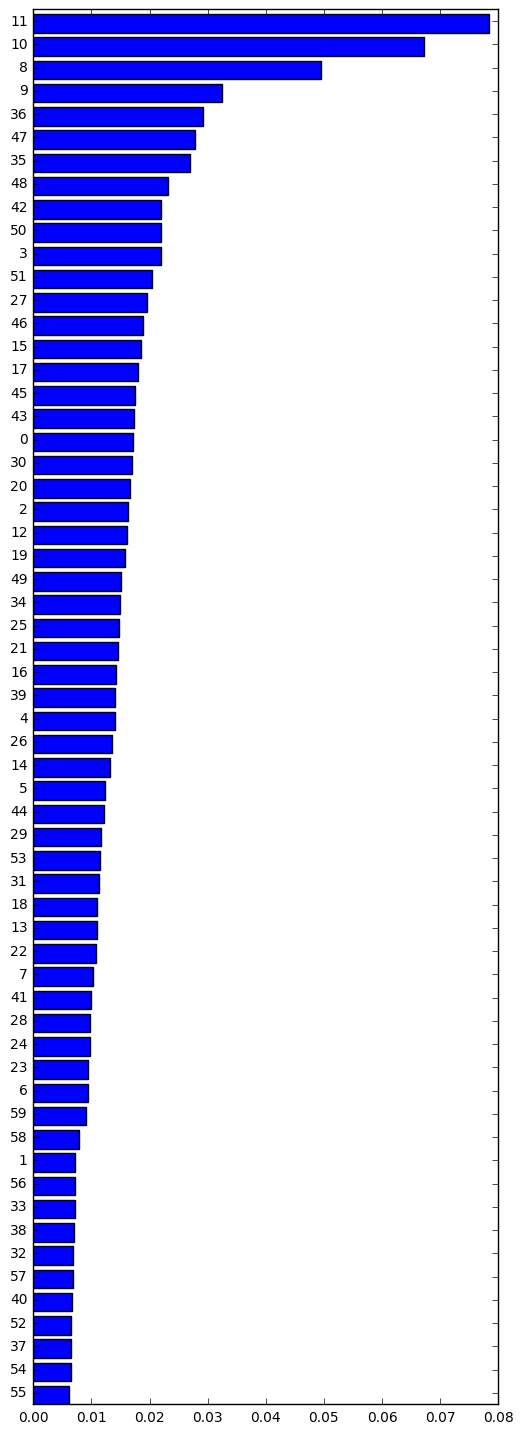
Which features are important?¶
imp = clf.best_estimator_.feature_importances_
idx = np.argsort(imp)
plt.figure(figsize=(6, 18))
plt.barh(range(len(imp)), imp[idx])
plt.yticks(np.arange(len(imp))+0.5, idx)
pass
Using a Pipeline¶
For cross-validation (e.g. grid search for best parameters), we often need to chain a series of steps and treat it as a single model. This chaining can be done with a Pipeline object.
from sklearn.pipeline import Pipeline
X_train, X_test, y_train, y_test = \
train_test_split(data_scaled, labels, test_size=0.33, random_state=42)
scaler = StandardScaler()
pca = PCA()
clf = LogisticRegression()
pipe = Pipeline(steps=[('scaler', scaler), ('pca', pca), ('clf', clf)])
n_components = [20, 30, 40, 50, 60]
Cs = np.logspace(-4, 4, 1)
estimator = GridSearchCV(pipe,
dict(pca__n_components=n_components,
clf__C=Cs), n_jobs=-1)
estimator.fit(X_train, y_train.codes)
pass
estimator.best_estimator_.named_steps['pca'].n_components
30
estimator.score(X_test, y_test.codes)
0.76811594202898548
y_true, y_pred = y_test.codes, estimator.predict(X_test)
print(classification_report(y_true, y_pred))
precision recall f1-score support
0 0.84 0.71 0.77 38
1 0.70 0.84 0.76 31
avg / total 0.78 0.77 0.77 69