from __future__ import division
import os
import sys
import glob
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
%matplotlib inline
%precision 4
plt.style.use('ggplot')
Fitting ODEs with the Levenberg–Marquardt algorithm¶
1D example¶
from lmfit import minimize, Parameters, Parameter, report_fit
from scipy.integrate import odeint
def f(xs, t, ps):
"""Receptor synthesis-internalization model."""
try:
a = ps['a'].value
b = ps['b'].value
except:
a, b = ps
x = xs
return a - b*x
def g(t, x0, ps):
"""
Solution to the ODE x'(t) = f(t,x,k) with initial condition x(0) = x0
"""
x = odeint(f, x0, t, args=(ps,))
return x
def residual(ps, ts, data):
x0 = ps['x0'].value
model = g(ts, x0, ps)
return (model - data).ravel()
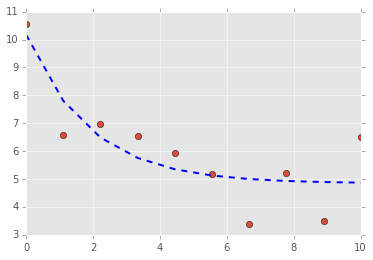
a = 2.0
b = 0.5
true_params = [a, b]
x0 = 10.0
t = np.linspace(0, 10, 10)
data = g(t, x0, true_params)
data += np.random.normal(size=data.shape)
# set parameters incluing bounds
params = Parameters()
params.add('x0', value=float(data[0]), min=0, max=100)
params.add('a', value= 1.0, min=0, max=10)
params.add('b', value= 1.0, min=0, max=10)
# fit model and find predicted values
result = minimize(residual, params, args=(t, data), method='leastsq')
final = data + result.residual.reshape(data.shape)
# plot data and fitted curves
plt.plot(t, data, 'o')
plt.plot(t, final, '--', linewidth=2, c='blue');
# display fitted statistics
report_fit(result)
[[Fit Statistics]]
# function evals = 29
# data points = 10
# variables = 3
chi-square = 10.080
reduced chi-square = 1.440
[[Variables]]
x0: 10.1714231 +/- 1.156777 (11.37%) (init= 10.54454)
a: 2.56428320 +/- 1.700899 (66.33%) (init= 1)
b: 0.52952597 +/- 0.296358 (55.97%) (init= 1)
[[Correlations]] (unreported correlations are < 0.100)
C(a, b) = 0.989
C(x0, b) = 0.453
C(x0, a) = 0.416
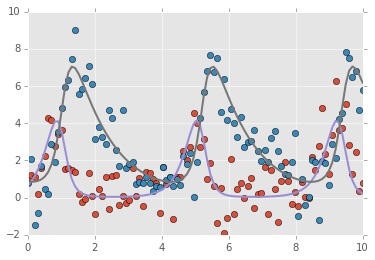
2D example¶
def f(xs, t, ps):
"""Lotka-Volterra predator-prey model."""
try:
a = ps['a'].value
b = ps['b'].value
c = ps['c'].value
d = ps['d'].value
except:
a, b, c, d = ps
x, y = xs
return [a*x - b*x*y, c*x*y - d*y]
def g(t, x0, ps):
"""
Solution to the ODE x'(t) = f(t,x,k) with initial condition x(0) = x0
"""
x = odeint(f, x0, t, args=(ps,))
return x
def residual(ps, ts, data):
x0 = ps['x0'].value, ps['y0'].value
model = g(ts, x0, ps)
return (model - data).ravel()
t = np.linspace(0, 10, 100)
x0 = np.array([1,1])
a, b, c, d = 3,1,1,1
true_params = np.array((a, b, c, d))
data = g(t, x0, true_params)
data += np.random.normal(size=data.shape)
# set parameters incluing bounds
params = Parameters()
params.add('x0', value= float(data[0, 0]), min=0, max=10)
params.add('y0', value=float(data[0, 1]), min=0, max=10)
params.add('a', value=2.0, min=0, max=10)
params.add('b', value=1.0, min=0, max=10)
params.add('c', value=1.0, min=0, max=10)
params.add('d', value=1.0, min=0, max=10)
# fit model and find predicted values
result = minimize(residual, params, args=(t, data), method='leastsq')
final = data + result.residual.reshape(data.shape)
# plot data and fitted curves
plt.plot(t, data, 'o')
plt.plot(t, final, '-', linewidth=2);
# display fitted statistics
report_fit(result)
[[Fit Statistics]]
# function evals = 106
# data points = 200
# variables = 6
chi-square = 195.573
reduced chi-square = 1.008
[[Variables]]
x0: 0.67757793 +/- 0.140751 (20.77%) (init= 0.9195372)
y0: 0.85617400 +/- 0.093697 (10.94%) (init= 0.7862886)
a: 3.72520718 +/- 0.423963 (11.38%) (init= 2)
b: 1.27267136 +/- 0.137525 (10.81%) (init= 1)
c: 1.03706693 +/- 0.087761 (8.46%) (init= 1)
d: 0.91828915 +/- 0.074839 (8.15%) (init= 1)
[[Correlations]] (unreported correlations are < 0.100)
C(a, b) = 0.953
C(a, d) = -0.926
C(x0, b) = -0.842
C(x0, a) = -0.829
C(b, d) = -0.822
C(y0, d) = -0.772
C(y0, c) = -0.686
C(x0, d) = 0.622
C(c, d) = 0.571
C(y0, a) = 0.516
C(a, c) = -0.374
C(y0, b) = 0.293
C(b, c) = -0.256
C(x0, y0) = -0.184