from __future__ import division
import os
import sys
import glob
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
%matplotlib inline
%precision 4
plt.style.use('ggplot')
np.random.seed(1234)
import pystan
import scipy.stats as stats
Using PyStan¶
Install PyStan with
pip install pystan
The nice thing about PyMC is that everything is in Python. With PyStan, however, you need to use a domain specific language based on C++ synteax to specify the model and the data, which is less flexible and more work. However, in exchange you get an extremely powerful HMC package (only does HMC) that can be used in R and Python.
References¶
Coin toss¶
We’ll repeat the example of determining the bias of a coin from observed coin tosses. The likelihood is binomial, and we use a beta prior.
coin_code = """
data {
int<lower=0> n; // number of tosses
int<lower=0> y; // number of heads
}
transformed data {}
parameters {
real<lower=0, upper=1> p;
}
transformed parameters {}
model {
p ~ beta(2, 2);
y ~ binomial(n, p);
}
generated quantities {}
"""
coin_dat = {
'n': 100,
'y': 61,
}
fit = pystan.stan(model_code=coin_code, data=coin_dat, iter=1000, chains=1)
Loading from a file¶
The string in coin_code can also be in a file - say coin_code.stan
- then we can use it like so
fit = pystan.stan(file='coin_code.stan', data=coin_dat, iter=1000, chains=1)
print(fit)
Inference for Stan model: anon_model_7f1947cd2d39ae427cd7b6bb6e6ffd77.
1 chains, each with iter=1000; warmup=500; thin=1;
post-warmup draws per chain=500, total post-warmup draws=500.
mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
p 0.61 4.9e-3 0.05 0.51 0.57 0.61 0.64 0.69 91.0 1.0
lp__ -70.22 0.06 0.66 -71.79 -70.43 -69.97 -69.79 -69.74 134.0 1.0
Samples were drawn using NUTS(diag_e) at Wed Mar 18 08:54:14 2015.
For each parameter, n_eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor on split chains (at
convergence, Rhat=1).
coin_dict = fit.extract()
coin_dict.keys()
# lp_ is the log posterior
[u'mu', u'sigma', u'lp__']
fit.plot('p');
plt.tight_layout()
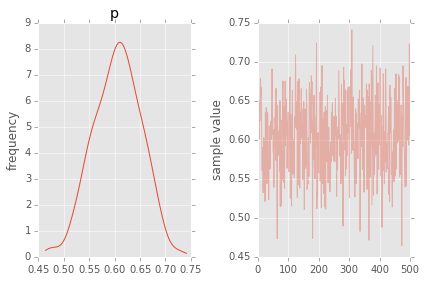
Estimating mean and standard deviation of normal distribution¶
norm_code = """
data {
int<lower=0> n;
real y[n];
}
transformed data {}
parameters {
real<lower=0, upper=100> mu;
real<lower=0, upper=10> sigma;
}
transformed parameters {}
model {
y ~ normal(mu, sigma);
}
generated quantities {}
"""
norm_dat = {
'n': 100,
'y': np.random.normal(10, 2, 100),
}
fit = pystan.stan(model_code=norm_code, data=norm_dat, iter=1000, chains=1)
print fit
Inference for Stan model: anon_model_3318343d5265d1b4ebc1e443f0228954.
1 chains, each with iter=1000; warmup=500; thin=1;
post-warmup draws per chain=500, total post-warmup draws=500.
mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
mu 10.09 0.02 0.19 9.72 9.97 10.09 10.22 10.49 120.0 1.0
sigma 2.02 0.01 0.15 1.74 1.92 2.01 2.12 2.32 119.0 1.01
lp__ -117.2 0.11 1.08 -120.0 -117.5 -116.8 -116.4 -116.2 105.0 1.0
Samples were drawn using NUTS(diag_e) at Wed Mar 18 08:54:50 2015.
For each parameter, n_eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor on split chains (at
convergence, Rhat=1).
trace = fit.extract()
plt.figure(figsize=(10,4))
plt.subplot(1,2,1);
plt.hist(trace['mu'][:], 25, histtype='step');
plt.subplot(1,2,2);
plt.hist(trace['sigma'][:], 25, histtype='step');
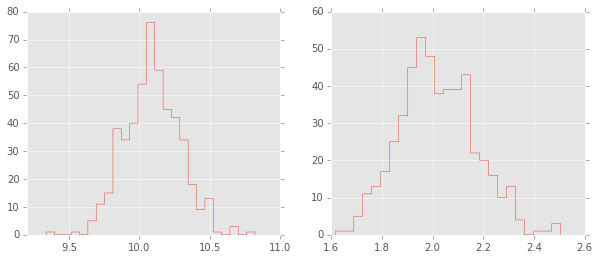
Optimization (finding MAP)¶
sm = pystan.StanModel(model_code=norm_code)
op = sm.optimizing(data=norm_dat)
op
OrderedDict([(u'mu', array(10.3016473417206)), (u'sigma', array(1.8823589782831152))])
Reusing fitted objects¶
new_dat = {
'n': 100,
'y': np.random.normal(10, 2, 100),
}
fit2 = pystan.stan(fit=fit, data=new_dat, chains=1)
print fit2
Inference for Stan model: anon_model_3318343d5265d1b4ebc1e443f0228954.
1 chains, each with iter=2000; warmup=1000; thin=1;
post-warmup draws per chain=1000, total post-warmup draws=1000.
mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
mu 9.89 0.01 0.19 9.54 9.76 9.9 10.02 10.27 250.0 1.0
sigma 1.99 9.3e-3 0.15 1.72 1.89 1.98 2.07 2.33 250.0 1.0
lp__ -115.4 0.08 1.01 -118.1 -115.8 -115.1 -114.7 -114.5 153.0 1.0
Samples were drawn using NUTS(diag_e) at Wed Mar 18 08:58:32 2015.
For each parameter, n_eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor on split chains (at
convergence, Rhat=1).
Saving compiled models¶
We can also compile Stan models and save them to file, so as to reload them for later use without needing to recompile.
def save(obj, filename):
"""Save compiled models for reuse."""
import pickle
with open(filename, 'w') as f:
pickle.dump(obj, f, protocol=pickle.HIGHEST_PROTOCOL)
def load(filename):
"""Reload compiled models for reuse."""
import pickle
return pickle.load(open(filename, 'r'))
model = pystan.StanModel(model_code=norm_code)
save(model, 'norm_model.pic')
new_model = load('norm_model.pic')
fit4 = new_model.sampling(new_dat, chains=1)
print fit4
Inference for Stan model: anon_model_3318343d5265d1b4ebc1e443f0228954.
1 chains, each with iter=2000; warmup=1000; thin=1;
post-warmup draws per chain=1000, total post-warmup draws=1000.
mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
mu 9.91 0.01 0.2 9.5 9.78 9.91 10.05 10.3 283.0 1.0
sigma 2.0 9.3e-3 0.15 1.73 1.9 1.99 2.09 2.31 244.0 1.0
lp__ -115.5 0.08 1.03 -118.2 -115.8 -115.1 -114.8 -114.5 153.0 1.01
Samples were drawn using NUTS(diag_e) at Wed Mar 18 09:18:30 2015.
For each parameter, n_eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor on split chains (at
convergence, Rhat=1).
Estimating parameters of a linear regreession model¶
We will show how to estimate regression parameters using a simple linear modesl
We can restate the linear model
as sampling from a probability distribution
We will assume the following priors
lin_reg_code = """
data {
int<lower=0> n;
real x[n];
real y[n];
}
transformed data {}
parameters {
real a;
real b;
real sigma;
}
transformed parameters {
real mu[n];
for (i in 1:n) {
mu[i] <- a*x[i] + b;
}
}
model {
sigma ~ uniform(0, 20);
y ~ normal(mu, sigma);
}
generated quantities {}
"""
n = 11
_a = 6
_b = 2
x = np.linspace(0, 1, n)
y = _a*x + _b + np.random.randn(n)
lin_reg_dat = {
'n': n,
'x': x,
'y': y
}
fit = pystan.stan(model_code=lin_reg_code, data=lin_reg_dat, iter=1000, chains=1)
print fit
fit.plot(['a', 'b']);
plt.tight_layout()
Simple Logistic model¶
We have observations of height and weight and want to use a logistic model to guess the sex.
# observed data
df = pd.read_csv('HtWt.csv')
df.head()
data {
int N; // number of obs (pregnancies)
int M; // number of groups (women)
int K; // number of predictors
int y[N]; // outcome
row_vector[K] x[N]; // predictors
int g[N]; // map obs to groups (pregnancies to women)
}
parameters {
real alpha;
real a[M];
vector[K] beta;
real sigma;
}
model {
sigma ~ uniform(0, 20);
a ~ normal(0, sigma);
b ~ normal(0,sigma);
c ~ normal(0, sigma)
for(n in 1:N) {
y[n] ~ bernoulli(inv_logit( alpha + a[g[n]] + x[n]*beta));
}
}'
log_reg_code = """
data {
int<lower=0> n;
int male[n];
real weight[n];
real height[n];
}
transformed data {}
parameters {
real a;
real b;
real c;
}
transformed parameters {}
model {
a ~ normal(0, 10);
b ~ normal(0, 10);
c ~ normal(0, 10);
for(i in 1:n) {
male[i] ~ bernoulli(inv_logit(a*weight[i] + b*height[i] + c));
}
}
generated quantities {}
"""
log_reg_dat = {
'n': len(df),
'male': df.male,
'height': df.height,
'weight': df.weight
}
fit = pystan.stan(model_code=log_reg_code, data=log_reg_dat, iter=2000, chains=1)
print fit
df_trace = pd.DataFrame(fit.extract(['c', 'b', 'a']))
pd.scatter_matrix(df_trace[:], diagonal='kde');
Estimating parameters of a logistic model¶
Gelman’s book has an example where the dose of a drug may be affected to the number of rat deaths in an experiment.
| Dose (log g/ml) | # Rats | # Deaths |
|---|---|---|
| -0.896 | 5 | 0 |
| -0.296 | 5 | 1 |
| -0.053 | 5 | 3 |
| 0.727 | 5 | 5 |
We will model the number of deaths as a random sample from a binomial distribution, where \(n\) is the number of rats and \(p\) the probabbility of a rat dying. We are given \(n = 5\), but we believve that \(p\) may be related to the drug dose \(x\). As \(x\) increases the number of rats dying seems to increase, and since \(p\) is a probability, we use the following model:
where we set vague priors for \(\alpha\) and \(\beta\), the parameters for the logistic model.
Original PyMC3 code¶
n = 5 * np.ones(4)
x = np.array([-0.896, -0.296, -0.053, 0.727])
y = np.array([0, 1, 3, 5])
def invlogit(x):
return pm.exp(x) / (1 + pm.exp(x))
with pm.Model() as model:
# define priors
alpha = pm.Normal('alpha', mu=0, sd=5)
beta = pm.Flat('beta')
# define likelihood
p = invlogit(alpha + beta*x)
y_obs = pm.Binomial('y_obs', n=n, p=p, observed=y)
# inference
start = pm.find_MAP()
step = pm.NUTS()
trace = pm.sample(niter, step, start, random_seed=123, progressbar=True)
Exercise - convert to PyStan version
Using a hierarchcical model¶
This uses the Gelman radon data set and is based off this IPython notebook. Radon levels were measured in houses from all counties in several states. Here we want to know if the preence of a basement affects the level of radon, and if this is affected by which county the house is located in.
The data set provided is just for the state of Minnesota, which has 85
counties with 2 to 116 measurements per county. We only need 3 columns
for this example county, log_radon, floor, where floor=0
indicates that there is a basement.
We will perfrom simple linear regression on log_radon as a function of county and floor.
radon = pd.read_csv('radon.csv')[['county', 'floor', 'log_radon']]
radon.head()
Hiearchical model¶
With a hierarchical model, there is an \(a_c\) and a \(b_c\) for each county \(c\) just as in the individual couty model, but they are no longer indepnedent but assumed to come from a common group distribution
we furhter assume that the hyperparameters come from the following distributions
Original PyMC3 code¶
county = pd.Categorical(radon['county']).codes
with pm.Model() as hm:
# County hyperpriors
mu_a = pm.Normal('mu_a', mu=0, tau=1.0/100**2)
sigma_a = pm.Uniform('sigma_a', lower=0, upper=100)
mu_b = pm.Normal('mu_b', mu=0, tau=1.0/100**2)
sigma_b = pm.Uniform('sigma_b', lower=0, upper=100)
# County slopes and intercepts
a = pm.Normal('slope', mu=mu_a, sd=sigma_a, shape=len(set(county)))
b = pm.Normal('intercept', mu=mu_b, tau=1.0/sigma_b**2, shape=len(set(county)))
# Houseehold errors
sigma = pm.Gamma("sigma", alpha=10, beta=1)
# Model prediction of radon level
mu = a[county] + b[county] * radon.floor.values
# Data likelihood
y = pm.Normal('y', mu=mu, sd=sigma, observed=radon.log_radon)
Exercise - convert to PyStan version