ML model examples¶
[1]:
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
sns.set_context('notebook', font_scale=1.5)
Dimension reduction¶
[2]:
from sklearn.datasets import load_breast_cancer
[3]:
bc = load_breast_cancer(as_frame=True)
[4]:
bc.data.head()
[4]:
| mean radius | mean texture | mean perimeter | mean area | mean smoothness | mean compactness | mean concavity | mean concave points | mean symmetry | mean fractal dimension | ... | worst radius | worst texture | worst perimeter | worst area | worst smoothness | worst compactness | worst concavity | worst concave points | worst symmetry | worst fractal dimension | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 17.99 | 10.38 | 122.80 | 1001.0 | 0.11840 | 0.27760 | 0.3001 | 0.14710 | 0.2419 | 0.07871 | ... | 25.38 | 17.33 | 184.60 | 2019.0 | 0.1622 | 0.6656 | 0.7119 | 0.2654 | 0.4601 | 0.11890 |
| 1 | 20.57 | 17.77 | 132.90 | 1326.0 | 0.08474 | 0.07864 | 0.0869 | 0.07017 | 0.1812 | 0.05667 | ... | 24.99 | 23.41 | 158.80 | 1956.0 | 0.1238 | 0.1866 | 0.2416 | 0.1860 | 0.2750 | 0.08902 |
| 2 | 19.69 | 21.25 | 130.00 | 1203.0 | 0.10960 | 0.15990 | 0.1974 | 0.12790 | 0.2069 | 0.05999 | ... | 23.57 | 25.53 | 152.50 | 1709.0 | 0.1444 | 0.4245 | 0.4504 | 0.2430 | 0.3613 | 0.08758 |
| 3 | 11.42 | 20.38 | 77.58 | 386.1 | 0.14250 | 0.28390 | 0.2414 | 0.10520 | 0.2597 | 0.09744 | ... | 14.91 | 26.50 | 98.87 | 567.7 | 0.2098 | 0.8663 | 0.6869 | 0.2575 | 0.6638 | 0.17300 |
| 4 | 20.29 | 14.34 | 135.10 | 1297.0 | 0.10030 | 0.13280 | 0.1980 | 0.10430 | 0.1809 | 0.05883 | ... | 22.54 | 16.67 | 152.20 | 1575.0 | 0.1374 | 0.2050 | 0.4000 | 0.1625 | 0.2364 | 0.07678 |
5 rows × 30 columns
[5]:
bc.target_names
[5]:
array(['malignant', 'benign'], dtype='<U9')
[6]:
bc.target.head()
[6]:
0 0
1 0
2 0
3 0
4 0
Name: target, dtype: int64
[7]:
! python3 -m pip install --quiet umap-learn
! python3 -m pip install --quiet phate
[8]:
from sklearn.decomposition import PCA
from sklearn.manifold import TSNE
from umap import UMAP
from phate import PHATE
[9]:
dr_models = {
'PCA': PCA(),
't-SNE': TSNE(),
'UMAP': UMAP(),
'PHATE': PHATE(verbose=0),
}
[10]:
from sklearn.preprocessing import StandardScaler
[11]:
scaler = StandardScaler()
[12]:
fig, axes = plt.subplots(2,2,figsize=(8,8))
axes = axes.ravel()
for i, (k, v) in enumerate(dr_models.items()):
X = v.fit_transform(scaler.fit_transform(bc.data))
target = bc.target
ax = axes[i]
ax.scatter(X[:, 0], X[:, 1], c=target)
ax.set_xlabel(f'{k}1')
ax.set_ylabel(f'{k}2')
ax.set_xticks([])
ax.set_yticks([])
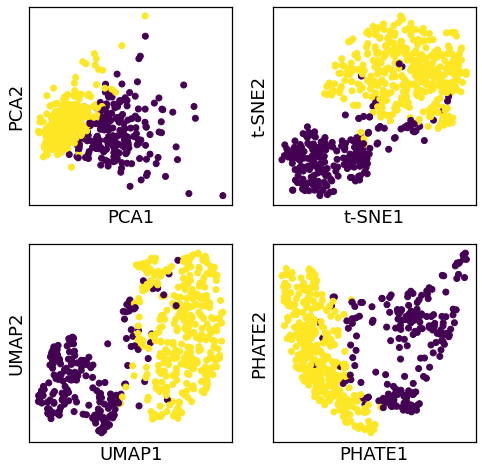
A3.2 Clustering¶
K-means
Agglomerative hierarchical clustering
Mixture models
[13]:
from sklearn.cluster import KMeans, AgglomerativeClustering, DBSCAN
from sklearn.mixture import GaussianMixture
[14]:
cl_models = {
'true': None,
'k-means': KMeans(n_clusters=2),
'ahc': AgglomerativeClustering(n_clusters=2),
'gmm': GaussianMixture(n_components=2),
}
[15]:
pca = PCA()
X = pca.fit_transform(scaler.fit_transform(bc.data))
[16]:
fig, axes = plt.subplots(2,2,figsize=(8, 8))
axes = axes.ravel()
for i, (k, v) in enumerate(cl_models.items()):
if i == 0:
y = bc.target
else:
y = v.fit_predict(scaler.fit_transform(bc.data))
target = y
ax = axes[i]
ax.scatter(X[:, 0], X[:, 1], c=target)
ax.set_xlabel('PC1')
ax.set_ylabel('PC2')
ax.set_xticks([])
ax.set_yticks([])
ax.set_title(k)
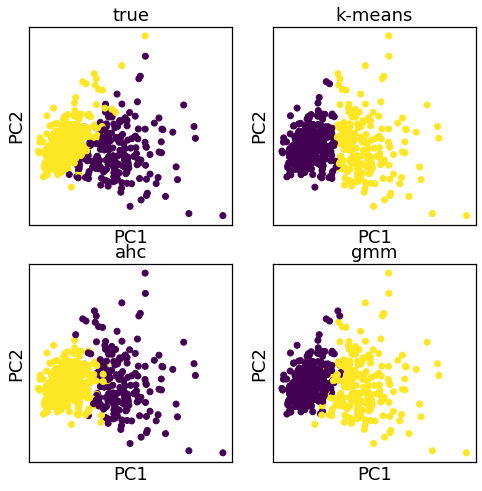
A3.3 Supervised learning¶
Nearest neighbor
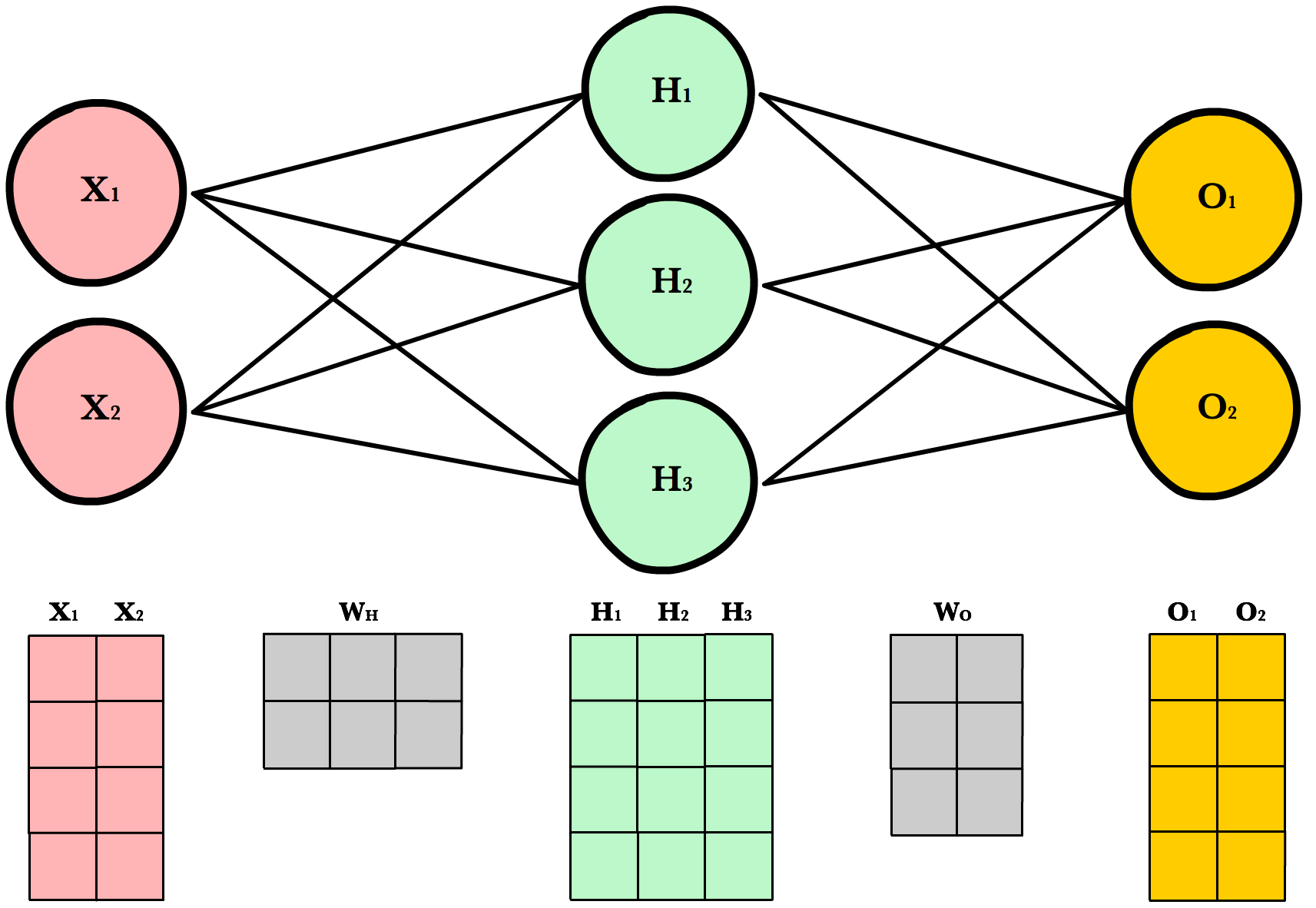
Linear models

Support vector machines

Trees

Neural networks

[17]:
from sklearn.model_selection import train_test_split
[18]:
from sklearn.dummy import DummyClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC
from sklearn.tree import DecisionTreeClassifier
from sklearn.neural_network import MLPClassifier
Proprocess data¶
[19]:
X = bc.data
y = bc.target
[20]:
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=0, stratify=y)
[21]:
X_train = scaler.fit_transform(X_train)
X_test = scaler.transform(X_test)
[22]:
pd.Series(y_test).value_counts(normalize=True)
[22]:
1 0.629371
0 0.370629
Name: target, dtype: float64
[23]:
sl_modles = dict(
dummy = DummyClassifier(strategy='prior'),
knn = KNeighborsClassifier(),
lr = LogisticRegression(),
svc = SVC(),
nn = MLPClassifier(max_iter=500),
)
[24]:
for name, clf in sl_modles.items():
clf.fit(X_train, y_train)
score = clf.score(X_test, y_test)
print(f'{name}: {score:.3f}')
dummy: 0.629
knn: 0.951
lr: 0.958
svc: 0.958
nn: 0.958