Hyperparameter tuning¶
There are 4 basic steps to hyperparameter tuning
Define the objective function
Define the space of hyperparameters to sample from
Define the metrics to optimize on
Run an optimization algorithm
The two simplest optimization algorithms are brute force search (aka Grid Search) and random sampling from the parameter space. Of course there are also more sophisticated search methods.
There are packages that provide wrappers for sklearn models and automatically use the model’s objective function, making the automation of tuning such models quite easy.
In practice, optimization is usually done over multiple model families - for simplicity we show only one model family but it is easy to find examples online.
[1]:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
Load data¶
We use squeeze to convert a single column DataFrame to a Series.
[2]:
X_train = pd.read_csv('data/X_train.csv')
X_test = pd.read_csv('data/X_test.csv')
y_train = pd.read_csv('data/y_train.csv').squeeze()
y_test = pd.read_csv('data/y_test.csv').squeeze()
Using skelearn¶
[3]:
from sklearn.ensemble import RandomForestClassifier
[4]:
RandomForestClassifier().get_params().keys()
[4]:
dict_keys(['bootstrap', 'ccp_alpha', 'class_weight', 'criterion', 'max_depth', 'max_features', 'max_leaf_nodes', 'max_samples', 'min_impurity_decrease', 'min_impurity_split', 'min_samples_leaf', 'min_samples_split', 'min_weight_fraction_leaf', 'n_estimators', 'n_jobs', 'oob_score', 'random_state', 'verbose', 'warm_start'])
You can run
help(RandomForestClassifier)
to get more information about the parameters.
For optimization, you need to understand the ML algorithm and what each tuning parameter does, and also to have some idea of what is a sensible range of values for each parameter. If in doubt, some orders of magnitude below and above the default value is a simple heuristic. In this example, we will tune the following (sklearn defaults in parentheses):
criterion= measure used to determine whether to split (Gini)n_estimators= n of trees (100)max_features= max number of features considered for splitting a node (sqrt(n_features)max_depth= max number of levels in each decision tree (None)min_samples_split= min number of data points placed in a node before the node is split (2)min_samples_leaf= min number of data points allowed in a leaf node (1)
We will search over the following
criterionGini, entropyn_estimators50, 100, 200max_features0.1, 0.3, 0.5,sqrt,logmax_depth1, 3, 5, 10, Nonemin_samples_split2, 5, 10min_samples_leaf1, 2, 5, 10
[5]:
from sklearn.model_selection import (
RandomizedSearchCV,
GridSearchCV
)
[6]:
params1 = {
'criterion': ['gini', 'entropy'],
'n_estimators': [50, 100, 200],
'max_features': [0.1, 0.3, 0.5, 'sqrt', 'log'],
'max_depth': [1, 3, 5, 10, None],
'min_samples_split': [2, 5, 10],
'min_samples_leaf': [1, 2, 5, 10],
}
This looks simple enough, but there are 1800 combinations to search!
[7]:
np.prod(list(map(len, params1.values())))
[7]:
1800
[8]:
rf = RandomForestClassifier()
Grid Search¶
[9]:
%%time
clf_gs = GridSearchCV(
rf,
params1,
n_jobs=-1,
).fit(X_train, y_train)
CPU times: user 20.9 s, sys: 1.79 s, total: 22.7 s
Wall time: 4min 44s
For other searchers, we limit to 32 trials simply to save time. In practice, you could use a larger number for a more comprehensive search.
[10]:
N = 32
Randomized Search¶
This is similar but searches a random subset of the specified parameters.
[11]:
%%time
clf_rs1 = RandomizedSearchCV(
rf,
params1,
n_jobs=-1,
random_state=0,
n_iter=N,
).fit(X_train, y_train)
CPU times: user 462 ms, sys: 20.9 ms, total: 482 ms
Wall time: 5.16 s
Using parameter distributions¶
[12]:
from scipy.stats import randint, uniform
[13]:
params2 = {
'criterion': ['gini', 'entropy'],
'n_estimators': randint(50, 201),
'max_features': uniform(0, 1),
'max_depth': randint(1, 101),
'min_samples_split': randint(2, 11),
'min_samples_leaf': randint(1, 11),
}
[14]:
%%time
clf_rs2 = RandomizedSearchCV(
rf,
params2,
n_jobs=-1,
random_state=0,
n_iter=N,
).fit(X_train, y_train)
CPU times: user 671 ms, sys: 34.4 ms, total: 705 ms
Wall time: 7.62 s
Using scikit-otpimize¶
[15]:
! python3 -m pip install --quiet scikit-optimize
[16]:
from skopt import BayesSearchCV
from skopt.space import Real, Categorical, Integer
[17]:
p = X_train.shape[1]
[18]:
params3 = {
'criterion': Categorical(['gini', 'entropy']),
'n_estimators': Integer(50, 201),
'max_features': Real(1/p, 1, prior='uniform'),
'max_depth': Integer(1, 101),
'min_samples_split': Integer(2, 11),
'min_samples_leaf': Integer(1, 11),
}
[19]:
%%time
clf_bs = BayesSearchCV(
rf,
params3,
n_jobs=-1,
n_iter=N,
random_state=0,
).fit(X_train, y_train)
CPU times: user 29.9 s, sys: 525 ms, total: 30.5 s
Wall time: 37.6 s
[20]:
from skopt.plots import plot_objective, plot_histogram
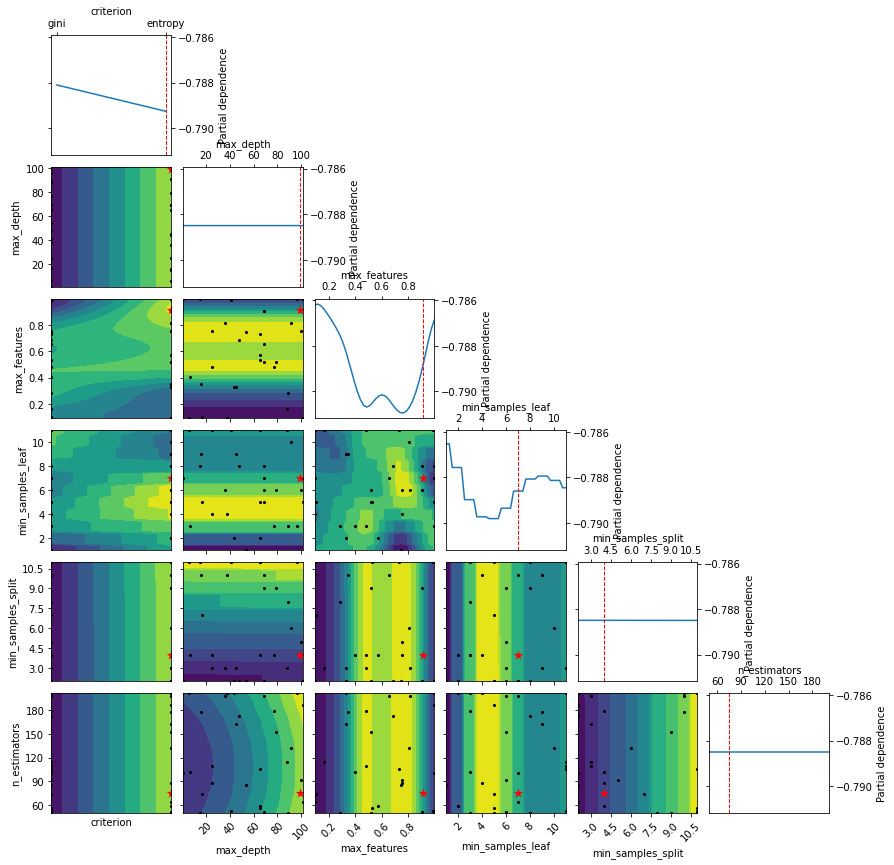
Show the partial dependency plot - an estimate of how features influence the objective function.
help(plot_objective)
[21]:
plot_objective(clf_bs.optimizer_results_[0],);
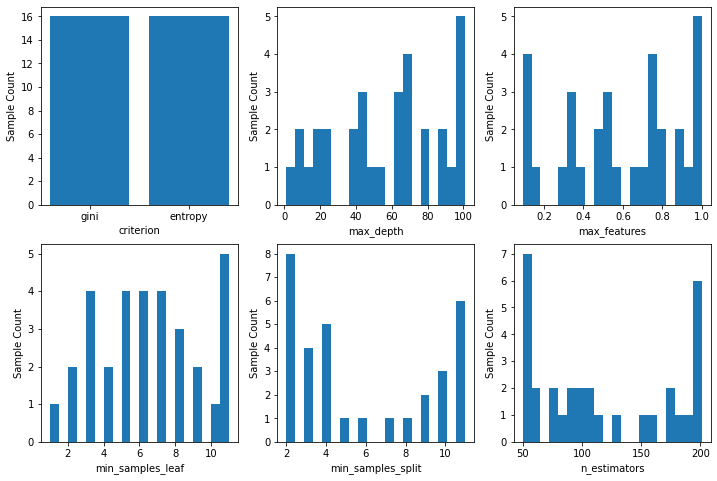
Show samples from the optimized space.
help(plot_histogram)
[22]:
fig, axes = plt.subplots(2,3,figsize=(12,8))
for i, ax in enumerate(axes.ravel()):
plot_histogram(clf_bs.optimizer_results_[0], i, ax=ax);
Using optuna¶
[23]:
! python3 -m pip install --quiet optuna
[24]:
import optuna
[25]:
from sklearn.model_selection import cross_val_score
[26]:
class Objective(object):
def __init__(self, X, y):
self.X = X
self.y = y
def __call__(self, trial):
X, y = self.X, self.y # load data once only
criterion = trial.suggest_categorical('criterion', ['gini', 'entropy'])
n_estimators = trial.suggest_int('n_estimators', 50, 201)
max_features = trial.suggest_float('max_features', 1/p, 1)
max_depth = trial.suggest_float('max_depth', 1, 128, log=True)
min_samples_split = trial.suggest_int('min_samples_split', 2, 11, 1)
min_samples_leaf = trial.suggest_int('min_samples_leaf', 1, 11, 1)
clf = RandomForestClassifier(
criterion = criterion,
n_estimators = n_estimators,
max_features = max_features,
max_depth = max_depth,
min_samples_split = min_samples_split,
min_samples_leaf = min_samples_leaf,
)
score = cross_val_score(clf, X, y, n_jobs=-1, cv=5).mean()
return score
[27]:
optuna.logging.set_verbosity(0)
[28]:
%%time
objective1 = Objective(X_train, y_train)
study1 = optuna.create_study(direction='maximize')
study1.optimize(objective1, n_trials=N)
CPU times: user 500 ms, sys: 18.3 ms, total: 519 ms
Wall time: 7.68 s
[29]:
df_results1 = study1.trials_dataframe()
[30]:
df_results1.head(3)
[30]:
| number | value | datetime_start | datetime_complete | duration | params_criterion | params_max_depth | params_max_features | params_min_samples_leaf | params_min_samples_split | params_n_estimators | state | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 0.788998 | 2020-10-12 15:56:31.009453 | 2020-10-12 15:56:31.302377 | 0 days 00:00:00.292924 | gini | 68.068025 | 0.317078 | 7 | 8 | 175 | COMPLETE |
| 1 | 1 | 0.767590 | 2020-10-12 15:56:31.302410 | 2020-10-12 15:56:31.390874 | 0 days 00:00:00.088464 | gini | 2.318150 | 0.477432 | 7 | 3 | 63 | COMPLETE |
| 2 | 2 | 0.762488 | 2020-10-12 15:56:31.390903 | 2020-10-12 15:56:31.480004 | 0 days 00:00:00.089101 | gini | 1.036565 | 0.974324 | 4 | 8 | 62 | COMPLETE |
[31]:
from sklearn.model_selection import train_test_split
[32]:
class ObjectiveES(object):
def __init__(self, X, y, max_iter=100):
self.X_train, self.X_val, self.y_train, self.y_val = train_test_split(X, y)
self.max_iter = max_iter
def __call__(self, trial):
# load ddta once only
X_train, y_train, X_val, y_val = self.X_train, self.y_train, self.X_val, self.y_val
criterion = trial.suggest_categorical('criterion', ['gini', 'entropy'])
n_estimators = trial.suggest_int('n_estimators', 50, 201)
max_features = trial.suggest_float('max_features', 1/p, 1)
max_depth = trial.suggest_float('max_depth', 1, 128, log=True)
min_samples_split = trial.suggest_int('min_samples_split', 2, 11, 1)
min_samples_leaf = trial.suggest_int('min_samples_leaf', 1, 11, 1)
clf = RandomForestClassifier(
criterion = criterion,
n_estimators = n_estimators,
max_features = max_features,
max_depth = max_depth,
min_samples_split = min_samples_split,
min_samples_leaf = min_samples_leaf,
)
for i in range(self.max_iter):
clf.fit(X_train, y_train)
score = clf.score(X_val, y_val)
trial.report(score, i)
if trial.should_prune():
raise optuna.TrialPruned()
return score
[33]:
%%time
objective2 = ObjectiveES(X_train, y_train)
study2 = optuna.create_study(direction='maximize')
study2.optimize(objective2, n_trials=N)
CPU times: user 6.63 s, sys: 24 ms, total: 6.65 s
Wall time: 6.66 s
[34]:
df_results2 = study2.trials_dataframe()
df_results2.head(3)
[34]:
| number | value | datetime_start | datetime_complete | duration | params_criterion | params_max_depth | params_max_features | params_min_samples_leaf | params_min_samples_split | params_n_estimators | state | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 0.813008 | 2020-10-12 15:56:38.736115 | 2020-10-12 15:56:38.915718 | 0 days 00:00:00.179603 | entropy | 10.027229 | 0.639109 | 11 | 9 | 87 | COMPLETE |
| 1 | 1 | 0.804878 | 2020-10-12 15:56:38.915750 | 2020-10-12 15:56:39.107483 | 0 days 00:00:00.191733 | entropy | 61.950795 | 0.343148 | 4 | 8 | 134 | COMPLETE |
| 2 | 2 | 0.841463 | 2020-10-12 15:56:39.107508 | 2020-10-12 15:56:39.359790 | 0 days 00:00:00.252282 | entropy | 60.831672 | 0.733779 | 2 | 7 | 152 | COMPLETE |
Visualizations¶
[35]:
from optuna.visualization import (plot_slice, plot_contour,
plot_optimization_history,
plot_param_importances,
plot_parallel_coordinate
)
[36]:
plot_optimization_history(study2)
[37]:
plot_param_importances(study2)
[38]:
plot_contour(
study2,
['min_samples_leaf', 'max_features', 'max_depth']
)
[39]:
plot_slice(study2, ['min_samples_leaf', 'max_features', 'max_depth'])
[40]:
plot_parallel_coordinate(study2, ['min_samples_leaf', 'max_features', 'max_depth'])
[41]:
clf_op1 = RandomForestClassifier(**study1.best_params)
clf_op1.fit(X_train, y_train)
clf_op2 = RandomForestClassifier(**study2.best_params)
clf_op2.fit(X_train, y_train);
[42]:
classifiers = [clf_gs, clf_rs1, clf_rs2, clf_bs, clf_op1, clf_op2]
names = ['Grid Search', 'Radnomized Search 1', 'Randomized Search 2',
'Bayesian', 'Optuna', 'Optuna Pruned']
[43]:
for name, clf in zip(names, classifiers):
print(f'{name:20s}: {clf.score(X_test, y_test): .3f}')
Grid Search : 0.848
Radnomized Search 1 : 0.826
Randomized Search 2 : 0.841
Bayesian : 0.829
Optuna : 0.829
Optuna Pruned : 0.823
Using pycaret¶
pycaret does not do anything that we have not done manually. However it presents a nice API that automates most of the boilerplate work in setting up a machine learning pipeline.
[44]:
! python3 -m pip install --quiet pycaret
[45]:
from pycaret.classification import (
setup,
compare_models,
plot_model,
create_model,
tune_model,
predict_model,
stack_models,
save_model,
load_model,
)
[46]:
data = X_train.copy()
data['survived'] = y_train
[47]:
clfs = setup(
data = data,
target = 'survived',
silent=True,
session_id=1,
)
Setup Succesfully Completed!
| Description | Value | |
|---|---|---|
| 0 | session_id | 1 |
| 1 | Target Type | Binary |
| 2 | Label Encoded | 0: 0, 1: 1 |
| 3 | Original Data | (981, 12) |
| 4 | Missing Values | False |
| 5 | Numeric Features | 5 |
| 6 | Categorical Features | 6 |
| 7 | Ordinal Features | False |
| 8 | High Cardinality Features | False |
| 9 | High Cardinality Method | None |
| 10 | Sampled Data | (981, 12) |
| 11 | Transformed Train Set | (686, 17) |
| 12 | Transformed Test Set | (295, 17) |
| 13 | Numeric Imputer | mean |
| 14 | Categorical Imputer | constant |
| 15 | Normalize | False |
| 16 | Normalize Method | None |
| 17 | Transformation | False |
| 18 | Transformation Method | None |
| 19 | PCA | False |
| 20 | PCA Method | None |
| 21 | PCA Components | None |
| 22 | Ignore Low Variance | False |
| 23 | Combine Rare Levels | False |
| 24 | Rare Level Threshold | None |
| 25 | Numeric Binning | False |
| 26 | Remove Outliers | False |
| 27 | Outliers Threshold | None |
| 28 | Remove Multicollinearity | False |
| 29 | Multicollinearity Threshold | None |
| 30 | Clustering | False |
| 31 | Clustering Iteration | None |
| 32 | Polynomial Features | False |
| 33 | Polynomial Degree | None |
| 34 | Trignometry Features | False |
| 35 | Polynomial Threshold | None |
| 36 | Group Features | False |
| 37 | Feature Selection | False |
| 38 | Features Selection Threshold | None |
| 39 | Feature Interaction | False |
| 40 | Feature Ratio | False |
| 41 | Interaction Threshold | None |
| 42 | Fix Imbalance | False |
| 43 | Fix Imbalance Method | SMOTE |
[48]:
best_model = compare_models(sort = 'Accuracy')
| Model | Accuracy | AUC | Recall | Prec. | F1 | Kappa | MCC | TT (Sec) | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | K Neighbors Classifier | 0.8001 | 0.8192 | 0.6925 | 0.7477 | 0.7178 | 0.5637 | 0.5656 | 0.0031 |
| 1 | CatBoost Classifier | 0.7942 | 0.8443 | 0.6572 | 0.7560 | 0.7019 | 0.5463 | 0.5503 | 1.6133 |
| 2 | Logistic Regression | 0.7914 | 0.8271 | 0.6734 | 0.7426 | 0.7034 | 0.5437 | 0.5476 | 0.0155 |
| 3 | Light Gradient Boosting Machine | 0.7798 | 0.8298 | 0.6575 | 0.7300 | 0.6894 | 0.5198 | 0.5237 | 0.0375 |
| 4 | Linear Discriminant Analysis | 0.7797 | 0.8278 | 0.6772 | 0.7166 | 0.6936 | 0.5224 | 0.5251 | 0.0044 |
| 5 | Gradient Boosting Classifier | 0.7783 | 0.8246 | 0.6460 | 0.7317 | 0.6845 | 0.5146 | 0.5186 | 0.0926 |
| 6 | Ridge Classifier | 0.7768 | 0.0000 | 0.6694 | 0.7143 | 0.6881 | 0.5152 | 0.5183 | 0.0050 |
| 7 | Extreme Gradient Boosting | 0.7696 | 0.8223 | 0.6608 | 0.7016 | 0.6786 | 0.4995 | 0.5017 | 0.0808 |
| 8 | Extra Trees Classifier | 0.7564 | 0.7992 | 0.6534 | 0.6841 | 0.6662 | 0.4749 | 0.4772 | 0.1365 |
| 9 | Random Forest Classifier | 0.7506 | 0.8142 | 0.6215 | 0.6800 | 0.6456 | 0.4547 | 0.4586 | 0.0233 |
| 10 | Ada Boost Classifier | 0.7491 | 0.7955 | 0.6612 | 0.6659 | 0.6623 | 0.4629 | 0.4641 | 0.0783 |
| 11 | SVM - Linear Kernel | 0.7389 | 0.0000 | 0.6434 | 0.6666 | 0.6260 | 0.4332 | 0.4472 | 0.0050 |
| 12 | Decision Tree Classifier | 0.7361 | 0.7222 | 0.6657 | 0.6426 | 0.6503 | 0.4394 | 0.4431 | 0.0040 |
| 13 | Naive Bayes | 0.6704 | 0.8114 | 0.1402 | 0.8308 | 0.1891 | 0.1338 | 0.2157 | 0.0032 |
| 14 | Quadratic Discriminant Analysis | 0.6298 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0028 |
[49]:
best_model
[49]:
KNeighborsClassifier(algorithm='auto', leaf_size=30, metric='minkowski',
metric_params=None, n_jobs=-1, n_neighbors=5, p=2,
weights='uniform')
[50]:
clf = create_model('gbc')
| Accuracy | AUC | Recall | Prec. | F1 | Kappa | MCC | |
|---|---|---|---|---|---|---|---|
| 0 | 0.7681 | 0.8524 | 0.6538 | 0.7083 | 0.6800 | 0.4986 | 0.4996 |
| 1 | 0.8551 | 0.8945 | 0.6923 | 0.9000 | 0.7826 | 0.6767 | 0.6898 |
| 2 | 0.6812 | 0.7276 | 0.5385 | 0.5833 | 0.5600 | 0.3106 | 0.3112 |
| 3 | 0.7971 | 0.9222 | 0.6154 | 0.8000 | 0.6957 | 0.5473 | 0.5579 |
| 4 | 0.8551 | 0.8955 | 0.7600 | 0.8261 | 0.7917 | 0.6809 | 0.6822 |
| 5 | 0.7971 | 0.7905 | 0.6400 | 0.7619 | 0.6957 | 0.5452 | 0.5499 |
| 6 | 0.7647 | 0.7907 | 0.6000 | 0.7143 | 0.6522 | 0.4764 | 0.4806 |
| 7 | 0.6912 | 0.7660 | 0.5600 | 0.5833 | 0.5714 | 0.3302 | 0.3304 |
| 8 | 0.8088 | 0.7870 | 0.6800 | 0.7727 | 0.7234 | 0.5782 | 0.5810 |
| 9 | 0.7647 | 0.8200 | 0.7200 | 0.6667 | 0.6923 | 0.5023 | 0.5033 |
| Mean | 0.7783 | 0.8246 | 0.6460 | 0.7317 | 0.6845 | 0.5146 | 0.5186 |
| SD | 0.0556 | 0.0606 | 0.0662 | 0.0967 | 0.0723 | 0.1172 | 0.1196 |
[51]:
tuned_clf = tune_model(clf)
| Accuracy | AUC | Recall | Prec. | F1 | Kappa | MCC | |
|---|---|---|---|---|---|---|---|
| 0 | 0.8116 | 0.8488 | 0.6154 | 0.8421 | 0.7111 | 0.5763 | 0.5919 |
| 1 | 0.8696 | 0.9244 | 0.6923 | 0.9474 | 0.8000 | 0.7067 | 0.7258 |
| 2 | 0.7101 | 0.7241 | 0.5000 | 0.6500 | 0.5652 | 0.3533 | 0.3602 |
| 3 | 0.8406 | 0.8927 | 0.6538 | 0.8947 | 0.7556 | 0.6415 | 0.6589 |
| 4 | 0.8696 | 0.8973 | 0.7200 | 0.9000 | 0.8000 | 0.7050 | 0.7147 |
| 5 | 0.8116 | 0.8250 | 0.6400 | 0.8000 | 0.7111 | 0.5739 | 0.5817 |
| 6 | 0.7500 | 0.7851 | 0.4800 | 0.7500 | 0.5854 | 0.4185 | 0.4399 |
| 7 | 0.7059 | 0.7651 | 0.5200 | 0.6190 | 0.5652 | 0.3455 | 0.3485 |
| 8 | 0.8088 | 0.8079 | 0.5600 | 0.8750 | 0.6829 | 0.5553 | 0.5837 |
| 9 | 0.7647 | 0.8400 | 0.6400 | 0.6957 | 0.6667 | 0.4853 | 0.4863 |
| Mean | 0.7942 | 0.8310 | 0.6022 | 0.7974 | 0.6843 | 0.5361 | 0.5492 |
| SD | 0.0566 | 0.0598 | 0.0785 | 0.1080 | 0.0847 | 0.1258 | 0.1291 |
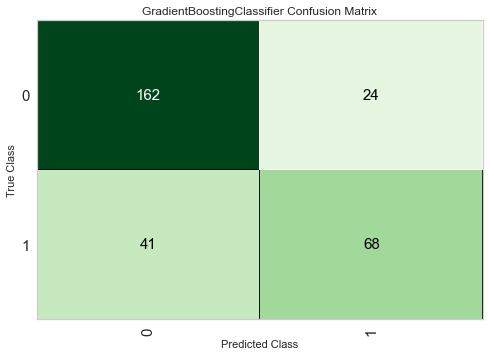
[52]:
plot_model(tuned_clf, 'confusion_matrix')
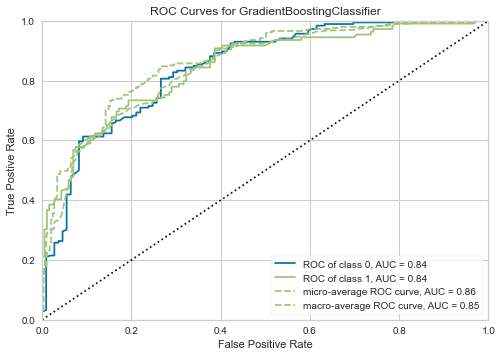
[53]:
plot_model(tuned_clf, 'auc')
[54]:
plot_model(tuned_clf, 'pr')
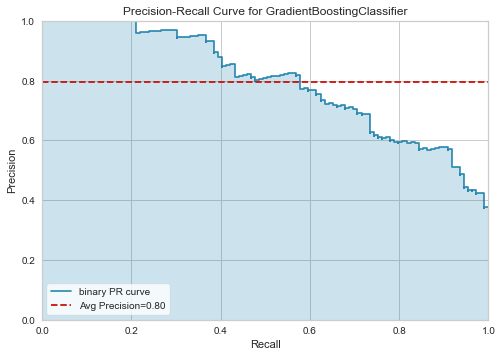
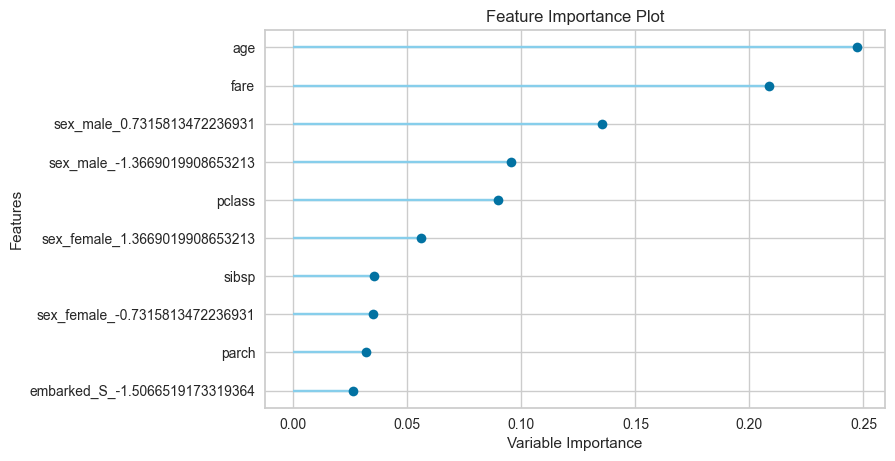
[55]:
plot_model(tuned_clf, 'feature')
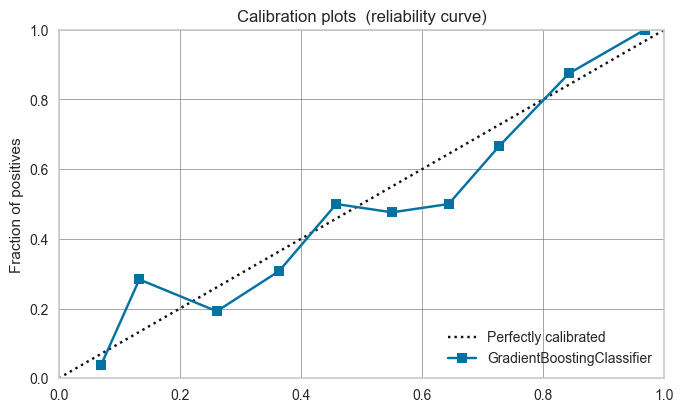
A calibration plot bins the test samples based on their predicted probabilities. If the predictions are good, the proportions should match the mean probability of the bin (i.e. be on the dotted line).
Models can be calibrated if the calibration plot shows a poor fit.
[56]:
plot_model(clf, plot='calibration')
[57]:
predict_model(clf);
| Model | Accuracy | AUC | Recall | Prec. | F1 | Kappa | MCC | |
|---|---|---|---|---|---|---|---|---|
| 0 | Gradient Boosting Classifier | 0.7932 | 0.8614 | 0.6514 | 0.7553 | 0.6995 | 0.5432 | 0.5467 |
[58]:
predict_model(tuned_clf);
| Model | Accuracy | AUC | Recall | Prec. | F1 | Kappa | MCC | |
|---|---|---|---|---|---|---|---|---|
| 0 | Gradient Boosting Classifier | 0.7797 | 0.8441 | 0.6239 | 0.7391 | 0.6766 | 0.5113 | 0.5156 |
We check a few other models.
[59]:
cb = create_model('catboost');
| Accuracy | AUC | Recall | Prec. | F1 | Kappa | MCC | |
|---|---|---|---|---|---|---|---|
| 0 | 0.8261 | 0.8953 | 0.7308 | 0.7917 | 0.7600 | 0.6240 | 0.6252 |
| 1 | 0.8696 | 0.9329 | 0.7308 | 0.9048 | 0.8085 | 0.7113 | 0.7206 |
| 2 | 0.6812 | 0.7379 | 0.5000 | 0.5909 | 0.5417 | 0.2998 | 0.3023 |
| 3 | 0.8551 | 0.9258 | 0.7308 | 0.8636 | 0.7917 | 0.6817 | 0.6873 |
| 4 | 0.8841 | 0.9245 | 0.8000 | 0.8696 | 0.8333 | 0.7447 | 0.7462 |
| 5 | 0.7971 | 0.8168 | 0.6800 | 0.7391 | 0.7083 | 0.5532 | 0.5543 |
| 6 | 0.7500 | 0.8042 | 0.5600 | 0.7000 | 0.6222 | 0.4388 | 0.4449 |
| 7 | 0.7353 | 0.7930 | 0.5200 | 0.6842 | 0.5909 | 0.4006 | 0.4088 |
| 8 | 0.7941 | 0.7814 | 0.6400 | 0.7619 | 0.6957 | 0.5419 | 0.5466 |
| 9 | 0.7500 | 0.8307 | 0.6800 | 0.6538 | 0.6667 | 0.4668 | 0.4670 |
| Mean | 0.7942 | 0.8443 | 0.6572 | 0.7560 | 0.7019 | 0.5463 | 0.5503 |
| SD | 0.0622 | 0.0663 | 0.0954 | 0.0970 | 0.0926 | 0.1382 | 0.1383 |
[60]:
predict_model(cb);
| Model | Accuracy | AUC | Recall | Prec. | F1 | Kappa | MCC | |
|---|---|---|---|---|---|---|---|---|
| 0 | CatBoost Classifier | 0.7831 | 0.8622 | 0.633 | 0.7419 | 0.6832 | 0.5198 | 0.5236 |
[61]:
tuned_cb = tune_model(cb)
| Accuracy | AUC | Recall | Prec. | F1 | Kappa | MCC | |
|---|---|---|---|---|---|---|---|
| 0 | 0.8261 | 0.8819 | 0.7692 | 0.7692 | 0.7692 | 0.6297 | 0.6297 |
| 1 | 0.8841 | 0.9231 | 0.8077 | 0.8750 | 0.8400 | 0.7493 | 0.7508 |
| 2 | 0.7101 | 0.7366 | 0.5769 | 0.6250 | 0.6000 | 0.3733 | 0.3740 |
| 3 | 0.8406 | 0.9168 | 0.6923 | 0.8571 | 0.7660 | 0.6471 | 0.6556 |
| 4 | 0.8696 | 0.9218 | 0.7600 | 0.8636 | 0.8085 | 0.7102 | 0.7136 |
| 5 | 0.7681 | 0.8150 | 0.6000 | 0.7143 | 0.6522 | 0.4802 | 0.4843 |
| 6 | 0.7794 | 0.7716 | 0.5600 | 0.7778 | 0.6512 | 0.4960 | 0.5104 |
| 7 | 0.7647 | 0.8033 | 0.6400 | 0.6957 | 0.6667 | 0.4853 | 0.4863 |
| 8 | 0.8088 | 0.8233 | 0.6400 | 0.8000 | 0.7111 | 0.5709 | 0.5788 |
| 9 | 0.7353 | 0.8149 | 0.6800 | 0.6296 | 0.6538 | 0.4401 | 0.4409 |
| Mean | 0.7987 | 0.8408 | 0.6726 | 0.7607 | 0.7119 | 0.5582 | 0.5624 |
| SD | 0.0541 | 0.0629 | 0.0806 | 0.0877 | 0.0756 | 0.1167 | 0.1169 |
[62]:
predict_model(tuned_cb);
| Model | Accuracy | AUC | Recall | Prec. | F1 | Kappa | MCC | |
|---|---|---|---|---|---|---|---|---|
| 0 | CatBoost Classifier | 0.7966 | 0.8636 | 0.6147 | 0.7882 | 0.6907 | 0.5426 | 0.552 |
[63]:
lr = create_model('lr');
| Accuracy | AUC | Recall | Prec. | F1 | Kappa | MCC | |
|---|---|---|---|---|---|---|---|
| 0 | 0.7971 | 0.8453 | 0.6154 | 0.8000 | 0.6957 | 0.5473 | 0.5579 |
| 1 | 0.8116 | 0.8962 | 0.7308 | 0.7600 | 0.7451 | 0.5958 | 0.5961 |
| 2 | 0.6957 | 0.7039 | 0.5000 | 0.6190 | 0.5532 | 0.3264 | 0.3306 |
| 3 | 0.8841 | 0.9275 | 0.8077 | 0.8750 | 0.8400 | 0.7493 | 0.7508 |
| 4 | 0.8551 | 0.8755 | 0.8000 | 0.8000 | 0.8000 | 0.6864 | 0.6864 |
| 5 | 0.8261 | 0.8077 | 0.6800 | 0.8095 | 0.7391 | 0.6102 | 0.6154 |
| 6 | 0.7647 | 0.7995 | 0.6000 | 0.7143 | 0.6522 | 0.4764 | 0.4806 |
| 7 | 0.7206 | 0.7614 | 0.6800 | 0.6071 | 0.6415 | 0.4138 | 0.4156 |
| 8 | 0.7647 | 0.7749 | 0.5600 | 0.7368 | 0.6364 | 0.4672 | 0.4768 |
| 9 | 0.7941 | 0.8791 | 0.7600 | 0.7037 | 0.7308 | 0.5645 | 0.5656 |
| Mean | 0.7914 | 0.8271 | 0.6734 | 0.7426 | 0.7034 | 0.5437 | 0.5476 |
| SD | 0.0547 | 0.0660 | 0.0983 | 0.0805 | 0.0808 | 0.1203 | 0.1191 |
[64]:
predict_model(lr);
| Model | Accuracy | AUC | Recall | Prec. | F1 | Kappa | MCC | |
|---|---|---|---|---|---|---|---|---|
| 0 | Logistic Regression | 0.7763 | 0.8417 | 0.6514 | 0.7172 | 0.6827 | 0.5105 | 0.5119 |
[65]:
tuned_lr = tune_model(lr)
| Accuracy | AUC | Recall | Prec. | F1 | Kappa | MCC | |
|---|---|---|---|---|---|---|---|
| 0 | 0.7681 | 0.8488 | 0.7692 | 0.6667 | 0.7143 | 0.5208 | 0.5246 |
| 1 | 0.8116 | 0.8945 | 0.8462 | 0.7097 | 0.7719 | 0.6135 | 0.6204 |
| 2 | 0.6812 | 0.7039 | 0.6538 | 0.5667 | 0.6071 | 0.3411 | 0.3436 |
| 3 | 0.8841 | 0.9249 | 0.8846 | 0.8214 | 0.8519 | 0.7568 | 0.7582 |
| 4 | 0.8116 | 0.8764 | 0.8000 | 0.7143 | 0.7547 | 0.6026 | 0.6051 |
| 5 | 0.7826 | 0.8041 | 0.6800 | 0.7083 | 0.6939 | 0.5254 | 0.5257 |
| 6 | 0.7353 | 0.8060 | 0.7200 | 0.6207 | 0.6667 | 0.4491 | 0.4525 |
| 7 | 0.6912 | 0.7633 | 0.7200 | 0.5625 | 0.6316 | 0.3726 | 0.3810 |
| 8 | 0.7647 | 0.7758 | 0.6400 | 0.6957 | 0.6667 | 0.4853 | 0.4863 |
| 9 | 0.7794 | 0.8772 | 0.8400 | 0.6562 | 0.7368 | 0.5518 | 0.5643 |
| Mean | 0.7710 | 0.8275 | 0.7554 | 0.6722 | 0.7096 | 0.5219 | 0.5262 |
| SD | 0.0566 | 0.0651 | 0.0813 | 0.0731 | 0.0689 | 0.1152 | 0.1148 |
[66]:
predict_model(tuned_lr);
| Model | Accuracy | AUC | Recall | Prec. | F1 | Kappa | MCC | |
|---|---|---|---|---|---|---|---|---|
| 0 | Logistic Regression | 0.7593 | 0.8422 | 0.7156 | 0.661 | 0.6872 | 0.4921 | 0.4932 |
[67]:
top = compare_models(n_select = 5)
| Model | Accuracy | AUC | Recall | Prec. | F1 | Kappa | MCC | TT (Sec) | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | K Neighbors Classifier | 0.8001 | 0.8192 | 0.6925 | 0.7477 | 0.7178 | 0.5637 | 0.5656 | 0.0030 |
| 1 | CatBoost Classifier | 0.7942 | 0.8443 | 0.6572 | 0.7560 | 0.7019 | 0.5463 | 0.5503 | 1.6453 |
| 2 | Logistic Regression | 0.7914 | 0.8271 | 0.6734 | 0.7426 | 0.7034 | 0.5437 | 0.5476 | 0.0146 |
| 3 | Light Gradient Boosting Machine | 0.7798 | 0.8298 | 0.6575 | 0.7300 | 0.6894 | 0.5198 | 0.5237 | 0.0358 |
| 4 | Linear Discriminant Analysis | 0.7797 | 0.8278 | 0.6772 | 0.7166 | 0.6936 | 0.5224 | 0.5251 | 0.0045 |
| 5 | Gradient Boosting Classifier | 0.7783 | 0.8246 | 0.6460 | 0.7317 | 0.6845 | 0.5146 | 0.5186 | 0.0944 |
| 6 | Ridge Classifier | 0.7768 | 0.0000 | 0.6694 | 0.7143 | 0.6881 | 0.5152 | 0.5183 | 0.0046 |
| 7 | Extreme Gradient Boosting | 0.7696 | 0.8223 | 0.6608 | 0.7016 | 0.6786 | 0.4995 | 0.5017 | 0.0822 |
| 8 | Extra Trees Classifier | 0.7564 | 0.7992 | 0.6534 | 0.6841 | 0.6662 | 0.4749 | 0.4772 | 0.1305 |
| 9 | Random Forest Classifier | 0.7506 | 0.8142 | 0.6215 | 0.6800 | 0.6456 | 0.4547 | 0.4586 | 0.0232 |
| 10 | Ada Boost Classifier | 0.7491 | 0.7955 | 0.6612 | 0.6659 | 0.6623 | 0.4629 | 0.4641 | 0.0779 |
| 11 | SVM - Linear Kernel | 0.7389 | 0.0000 | 0.6434 | 0.6666 | 0.6260 | 0.4332 | 0.4472 | 0.0050 |
| 12 | Decision Tree Classifier | 0.7361 | 0.7222 | 0.6657 | 0.6426 | 0.6503 | 0.4394 | 0.4431 | 0.0040 |
| 13 | Naive Bayes | 0.6704 | 0.8114 | 0.1402 | 0.8308 | 0.1891 | 0.1338 | 0.2157 | 0.0034 |
| 14 | Quadratic Discriminant Analysis | 0.6298 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0033 |
[68]:
stack_clf = stack_models(top)
| Accuracy | AUC | Recall | Prec. | F1 | Kappa | MCC | |
|---|---|---|---|---|---|---|---|
| 0 | 0.8406 | 0.8801 | 0.7308 | 0.8261 | 0.7755 | 0.6526 | 0.6556 |
| 1 | 0.8696 | 0.9222 | 0.7308 | 0.9048 | 0.8085 | 0.7113 | 0.7206 |
| 2 | 0.7101 | 0.7030 | 0.5769 | 0.6250 | 0.6000 | 0.3733 | 0.3740 |
| 3 | 0.8696 | 0.9222 | 0.7692 | 0.8696 | 0.8163 | 0.7158 | 0.7190 |
| 4 | 0.8841 | 0.9009 | 0.8000 | 0.8696 | 0.8333 | 0.7447 | 0.7462 |
| 5 | 0.7826 | 0.8186 | 0.6400 | 0.7273 | 0.6809 | 0.5170 | 0.5195 |
| 6 | 0.7500 | 0.8312 | 0.6000 | 0.6818 | 0.6383 | 0.4485 | 0.4506 |
| 7 | 0.7647 | 0.7763 | 0.6000 | 0.7143 | 0.6522 | 0.4764 | 0.4806 |
| 8 | 0.8088 | 0.7842 | 0.6400 | 0.8000 | 0.7111 | 0.5709 | 0.5788 |
| 9 | 0.7794 | 0.8586 | 0.7200 | 0.6923 | 0.7059 | 0.5295 | 0.5298 |
| Mean | 0.8059 | 0.8397 | 0.6808 | 0.7711 | 0.7222 | 0.5740 | 0.5775 |
| SD | 0.0554 | 0.0676 | 0.0747 | 0.0904 | 0.0778 | 0.1204 | 0.1215 |
[69]:
predict_model(stack_clf);
| Model | Accuracy | AUC | Recall | Prec. | F1 | Kappa | MCC | |
|---|---|---|---|---|---|---|---|---|
| 0 | Stacking Classifier | 0.7898 | 0.8634 | 0.6514 | 0.7474 | 0.6961 | 0.5366 | 0.5396 |
[70]:
import pendulum
[71]:
today = pendulum.today()
[72]:
save_model(tuned_cb, f'Titanic Tuned CatBoost {today}')
Transformation Pipeline and Model Succesfully Saved
[73]:
!ls T*
Titanic Tuned CatBoost 2020-10-12T00:00:00-04:00.pkl
[74]:
clf = load_model(f'Titanic Tuned CatBoost {today}')
Transformation Pipeline and Model Successfully Loaded
[75]:
predict_model(tuned_cb);
| Model | Accuracy | AUC | Recall | Prec. | F1 | Kappa | MCC | |
|---|---|---|---|---|---|---|---|---|
| 0 | CatBoost Classifier | 0.7966 | 0.8636 | 0.6147 | 0.7882 | 0.6907 | 0.5426 | 0.552 |
[76]:
predict_model(clf);
| Model | Accuracy | AUC | Recall | Prec. | F1 | Kappa | MCC | |
|---|---|---|---|---|---|---|---|---|
| 0 | CatBoost Classifier | 0.7966 | 0.8636 | 0.6147 | 0.7882 | 0.6907 | 0.5426 | 0.552 |
[ ]: