Spark MLLib¶
The MLLib library has two packages - pyspark.mllib which provides an RDD interface, and pyspark.ml which provides a DataFrame interface. we will focus only on the high-level interface.
Topics:
Preparing a DataFrame for ML
Generating simple statistics
Splitting data
Preprocessing
Feature extraction
Imputation
Transforms
Unsupervised learning
Supervised learning
Hyperparameter optimization
Using R formula
Evaluation
Using pipelines
Set up Spark and Spark SQL contexts¶
[4]:
from pyspark.sql import SparkSession
[5]:
spark = (
SparkSession.builder
.master("local")
.appName("BIOS-823")
.config("spark.executor.cores", 4)
.getOrCreate()
)
Preparing a DataFrame for ML¶
Spark provides dense and sparse vectors and matrices mainly as data structures for ML, so even though they live in the ml.linalg module, the ability to manipulate them is very limited.
[6]:
import numpy as np
[7]:
from pyspark.ml.linalg import Vectors
Vectors¶
[8]:
v1 = Vectors.dense(range(1,11))
v2 = Vectors.dense(range(0, 10))
[9]:
v1
[9]:
DenseVector([1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0, 9.0, 10.0])
[10]:
v2
[10]:
DenseVector([0.0, 1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0, 9.0])
[11]:
v1.array
[11]:
array([ 1., 2., 3., 4., 5., 6., 7., 8., 9., 10.])
[12]:
v1.norm(p=1)
[12]:
55.0
[13]:
v1.sum()
[13]:
55.0
[14]:
v1.norm(p=2)
[14]:
19.621416870348583
[15]:
np.sqrt((v1*v1).sum())
[15]:
19.621416870348583
[16]:
v1.squared_distance(v2)
[16]:
10.0
[17]:
((v1 - v2).array**2).sum()
[17]:
10.0
[18]:
v1.dot(v2)
[18]:
330.0
Manual construction of an ML DaataFrame¶
ML operations generally require a DataFrame with columns corresponding to a a label and features represented as a Vector.
[19]:
data =[
(0.0, Vectors.dense([1,2,3])),
(1.0, Vectors.dense([2,3,4])),
(0.0, Vectors.dense([3,4,5])),
(1.0, Vectors.dense([2,2,2]))
]
df = spark.createDataFrame(data, ['label', 'features'])
[20]:
df.show()
+-----+-------------+
|label| features|
+-----+-------------+
| 0.0|[1.0,2.0,3.0]|
| 1.0|[2.0,3.0,4.0]|
| 0.0|[3.0,4.0,5.0]|
| 1.0|[2.0,2.0,2.0]|
+-----+-------------+
[21]:
from pyspark.ml.functions import vector_to_array
[22]:
from pyspark.sql.functions import col
[23]:
df.select('label', vector_to_array('features')).show()
+-----+---------------+
|label| UDF(features)|
+-----+---------------+
| 0.0|[1.0, 2.0, 3.0]|
| 1.0|[2.0, 3.0, 4.0]|
| 0.0|[3.0, 4.0, 5.0]|
| 1.0|[2.0, 2.0, 2.0]|
+-----+---------------+
[24]:
df.withColumn("xs", vector_to_array("features")).show()
+-----+-------------+---------------+
|label| features| xs|
+-----+-------------+---------------+
| 0.0|[1.0,2.0,3.0]|[1.0, 2.0, 3.0]|
| 1.0|[2.0,3.0,4.0]|[2.0, 3.0, 4.0]|
| 0.0|[3.0,4.0,5.0]|[3.0, 4.0, 5.0]|
| 1.0|[2.0,2.0,2.0]|[2.0, 2.0, 2.0]|
+-----+-------------+---------------+
[25]:
df.withColumn("xs", vector_to_array("features")).dtypes
[25]:
[('label', 'double'), ('features', 'vector'), ('xs', 'array<double>')]
[26]:
(
df.
withColumn("xs", vector_to_array("features")).
select(["label"] + [col("xs")[i] for i in range(3)])
).show()
+-----+-----+-----+-----+
|label|xs[0]|xs[1]|xs[2]|
+-----+-----+-----+-----+
| 0.0| 1.0| 2.0| 3.0|
| 1.0| 2.0| 3.0| 4.0|
| 0.0| 3.0| 4.0| 5.0|
| 1.0| 2.0| 2.0| 2.0|
+-----+-----+-----+-----+
Using VectorAssembler¶
[27]:
from pyspark.ml.feature import VectorAssembler
[28]:
import pandas as pd
[29]:
url = 'https://bit.ly/3eoBK6t'
pdf = pd.read_csv(url)
[30]:
pdf.head(3)
[30]:
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
[31]:
iris = spark.createDataFrame(pdf)
[32]:
iris.show(3)
+------------+-----------+------------+-----------+-------+
|sepal_length|sepal_width|petal_length|petal_width|species|
+------------+-----------+------------+-----------+-------+
| 5.1| 3.5| 1.4| 0.2| setosa|
| 4.9| 3.0| 1.4| 0.2| setosa|
| 4.7| 3.2| 1.3| 0.2| setosa|
+------------+-----------+------------+-----------+-------+
only showing top 3 rows
[33]:
iris.columns[:-1]
[33]:
['sepal_length', 'sepal_width', 'petal_length', 'petal_width']
[34]:
assembler = VectorAssembler(
inputCols=iris.columns[:-1],
outputCol='raw_features')
df = assembler.transform(iris)
[35]:
df.show(3)
+------------+-----------+------------+-----------+-------+-----------------+
|sepal_length|sepal_width|petal_length|petal_width|species| raw_features|
+------------+-----------+------------+-----------+-------+-----------------+
| 5.1| 3.5| 1.4| 0.2| setosa|[5.1,3.5,1.4,0.2]|
| 4.9| 3.0| 1.4| 0.2| setosa|[4.9,3.0,1.4,0.2]|
| 4.7| 3.2| 1.3| 0.2| setosa|[4.7,3.2,1.3,0.2]|
+------------+-----------+------------+-----------+-------+-----------------+
only showing top 3 rows
Generating simple statistics¶
[36]:
import pyspark.ml.stat as stat
[37]:
df.select(stat.Summarizer.mean(col('raw_features'))).show()
+--------------------+
| mean(raw_features)|
+--------------------+
|[5.84333333333333...|
+--------------------+
[38]:
(
df.select(
stat.Summarizer.metrics('std', 'min', 'max', 'mean').
summary(col('raw_features'))).
show(truncate=False)
)
+---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+
|aggregate_metrics(raw_features, 1.0) |
+---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+
|[[0.8280661279778628,0.43359431136217363,1.7644204199522624,0.7631607417008414], [4.3,2.0,1.0,0.1], [7.9,4.4,6.9,2.5], [5.843333333333335,3.0540000000000003,3.758666666666667,1.1986666666666668]]|
+---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+
Split data¶
[39]:
train_data, test_data = df.randomSplit(weights=[2.,1.], seed=0)
[40]:
train_data.count(), test_data.count()
[40]:
(98, 52)
[41]:
train_data.sample(0.1).show()
+------------+-----------+------------+-----------+----------+-----------------+
|sepal_length|sepal_width|petal_length|petal_width| species| raw_features|
+------------+-----------+------------+-----------+----------+-----------------+
| 5.0| 3.4| 1.5| 0.2| setosa|[5.0,3.4,1.5,0.2]|
| 5.4| 3.7| 1.5| 0.2| setosa|[5.4,3.7,1.5,0.2]|
| 4.8| 3.0| 1.4| 0.1| setosa|[4.8,3.0,1.4,0.1]|
| 4.8| 3.4| 1.9| 0.2| setosa|[4.8,3.4,1.9,0.2]|
| 4.9| 3.1| 1.5| 0.1| setosa|[4.9,3.1,1.5,0.1]|
| 5.2| 4.1| 1.5| 0.1| setosa|[5.2,4.1,1.5,0.1]|
| 5.4| 3.4| 1.5| 0.4| setosa|[5.4,3.4,1.5,0.4]|
| 5.6| 3.0| 4.5| 1.5|versicolor|[5.6,3.0,4.5,1.5]|
| 5.8| 2.7| 4.1| 1.0|versicolor|[5.8,2.7,4.1,1.0]|
| 6.1| 2.9| 4.7| 1.4|versicolor|[6.1,2.9,4.7,1.4]|
| 5.0| 2.3| 3.3| 1.0|versicolor|[5.0,2.3,3.3,1.0]|
| 5.5| 2.6| 4.4| 1.2|versicolor|[5.5,2.6,4.4,1.2]|
| 5.7| 3.0| 4.2| 1.2|versicolor|[5.7,3.0,4.2,1.2]|
| 6.4| 2.7| 5.3| 1.9| virginica|[6.4,2.7,5.3,1.9]|
| 6.5| 3.2| 5.1| 2.0| virginica|[6.5,3.2,5.1,2.0]|
| 6.7| 3.3| 5.7| 2.1| virginica|[6.7,3.3,5.7,2.1]|
| 6.4| 3.1| 5.5| 1.8| virginica|[6.4,3.1,5.5,1.8]|
| 6.9| 3.1| 5.1| 2.3| virginica|[6.9,3.1,5.1,2.3]|
+------------+-----------+------------+-----------+----------+-----------------+
[42]:
test_data.sample(0.1).show()
+------------+-----------+------------+-----------+----------+-----------------+
|sepal_length|sepal_width|petal_length|petal_width| species| raw_features|
+------------+-----------+------------+-----------+----------+-----------------+
| 5.0| 3.6| 1.4| 0.2| setosa|[5.0,3.6,1.4,0.2]|
| 5.2| 3.5| 1.5| 0.2| setosa|[5.2,3.5,1.5,0.2]|
| 4.9| 2.4| 3.3| 1.0|versicolor|[4.9,2.4,3.3,1.0]|
| 6.3| 2.9| 5.6| 1.8| virginica|[6.3,2.9,5.6,1.8]|
| 7.2| 3.2| 6.0| 1.8| virginica|[7.2,3.2,6.0,1.8]|
| 6.7| 3.3| 5.7| 2.5| virginica|[6.7,3.3,5.7,2.5]|
+------------+-----------+------------+-----------+----------+-----------------+
[43]:
train_data = train_data.persist()
Processing data¶
Encoding categorical features¶
[44]:
from pyspark.ml.feature import StringIndexer
[45]:
indexer = StringIndexer(inputCol='species', outputCol='label')
[46]:
indexer = indexer.fit(train_data)
[47]:
train_df = indexer.transform(train_data)
test_df = indexer.transform(test_data)
[48]:
train_df.sample(0.1).show()
+------------+-----------+------------+-----------+----------+-----------------+-----+
|sepal_length|sepal_width|petal_length|petal_width| species| raw_features|label|
+------------+-----------+------------+-----------+----------+-----------------+-----+
| 4.6| 3.4| 1.4| 0.3| setosa|[4.6,3.4,1.4,0.3]| 0.0|
| 4.9| 3.0| 1.4| 0.2| setosa|[4.9,3.0,1.4,0.2]| 0.0|
| 4.7| 3.2| 1.6| 0.2| setosa|[4.7,3.2,1.6,0.2]| 0.0|
| 6.6| 2.9| 4.6| 1.3|versicolor|[6.6,2.9,4.6,1.3]| 1.0|
| 5.6| 2.5| 3.9| 1.1|versicolor|[5.6,2.5,3.9,1.1]| 1.0|
| 5.9| 3.0| 4.2| 1.5|versicolor|[5.9,3.0,4.2,1.5]| 1.0|
| 6.2| 2.2| 4.5| 1.5|versicolor|[6.2,2.2,4.5,1.5]| 1.0|
| 6.7| 3.1| 4.4| 1.4|versicolor|[6.7,3.1,4.4,1.4]| 1.0|
| 6.4| 2.9| 4.3| 1.3|versicolor|[6.4,2.9,4.3,1.3]| 1.0|
| 5.5| 2.6| 4.4| 1.2|versicolor|[5.5,2.6,4.4,1.2]| 1.0|
| 6.2| 3.4| 5.4| 2.3| virginica|[6.2,3.4,5.4,2.3]| 2.0|
| 6.9| 3.1| 5.1| 2.3| virginica|[6.9,3.1,5.1,2.3]| 2.0|
+------------+-----------+------------+-----------+----------+-----------------+-----+
Scaling¶
[49]:
from pyspark.ml.feature import StandardScaler
[50]:
scaler = StandardScaler(withMean=True,
withStd=True,
inputCol='raw_features',
outputCol='features')
[51]:
scaler = scaler.fit(train_df)
[52]:
train_df_scaled = scaler.transform(train_df)
test_df_scaled = scaler.transform(test_df)
[53]:
train_df_scaled.select('features', 'features').show(5, truncate=False)
+---------------------------------------------------------------------------------+---------------------------------------------------------------------------------+
|features |features |
+---------------------------------------------------------------------------------+---------------------------------------------------------------------------------+
|[-1.5213158492369616,0.00692241917037384,-1.2132855899947563,-1.22045281288949] |[-1.5213158492369616,0.00692241917037384,-1.2132855899947563,-1.22045281288949] |
|[-1.5213158492369616,0.6853194978669278,-1.2714283713245784,-1.0882932817699649] |[-1.5213158492369616,0.6853194978669278,-1.2714283713245784,-1.0882932817699649] |
|[-1.390075573510533,0.23305477873589214,-1.3295711526544005,-1.22045281288949] |[-1.390075573510533,0.23305477873589214,-1.3295711526544005,-1.22045281288949] |
|[-1.1275950220576765,-0.21920994039514446,-1.2714283713245784,-1.22045281288949] |[-1.1275950220576765,-0.21920994039514446,-1.2714283713245784,-1.22045281288949] |
|[-1.1275950220576765,0.00692241917037384,-1.2132855899947563,-1.3526123440090148]|[-1.1275950220576765,0.00692241917037384,-1.2132855899947563,-1.3526123440090148]|
+---------------------------------------------------------------------------------+---------------------------------------------------------------------------------+
only showing top 5 rows
Unsupervised learning¶
Dimension reduction¶
[54]:
from pyspark.ml.feature import PCA
[55]:
pca = PCA(k=2, inputCol='features', outputCol='pc').fit(train_df_scaled)
[56]:
train_df_scaled = pca.transform(train_df_scaled)
[57]:
train_df_scaled.select('pc').show(5, truncate=False)
+----------------------------------------+
|pc |
+----------------------------------------+
|[2.1719561815281754,0.7342240484007609] |
|[2.3257684348824283,0.11499107410380768]|
|[2.2379887825335114,0.47537636776842684]|
|[1.9410238356438345,0.7700780683206433] |
|[2.0469675488421304,0.5739261085291683] |
+----------------------------------------+
only showing top 5 rows
[58]:
import matplotlib.pyplot as plt
[59]:
pdf = (
train_df_scaled.select('label', 'pc').
withColumn("pc", vector_to_array("pc")).
select(["label"] + [col("pc")[i] for i in range(2)])
).toPandas()
[60]:
pdf.head(3)
[60]:
| label | pc[0] | pc[1] | |
|---|---|---|---|
| 0 | 0.0 | 2.171956 | 0.734224 |
| 1 | 0.0 | 2.325768 | 0.114991 |
| 2 | 0.0 | 2.237989 | 0.475376 |
[61]:
plt.scatter(pdf.iloc[:, 1], pdf.iloc[:, 2], c=pdf.iloc[:, 0])
plt.xlabel('PC1')
plt.ylabel('PC2')
plt.tight_layout()
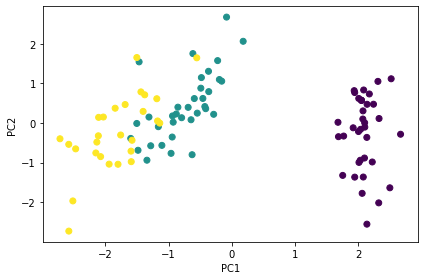
Clustering¶
[62]:
from pyspark.ml.clustering import GaussianMixture
[63]:
gmm = GaussianMixture(
featuresCol='features',
k=3,
seed=0)
[64]:
gmm = gmm.fit(train_df_scaled)
[65]:
gmm.gaussiansDF.toPandas()
[65]:
| mean | cov | |
|---|---|---|
| 0 | [-0.9380257346418976, 0.8046671327476248, -1.2... | DenseMatrix([[0.2166823 , 0.29970786, 0.013235... |
| 1 | [0.005938775919945091, -0.5221004264901148, 0.... | DenseMatrix([[ 0.10356088, -0.0849359 , 0.056... |
| 2 | [0.6061006081018865, -0.4609677242002642, 0.72... | DenseMatrix([[0.65443637, 0.40398667, 0.255851... |
[66]:
train_df_scaled = gmm.transform(train_df_scaled)
[67]:
train_df_scaled.select('features', 'label', 'probability', 'prediction').sample(0.1).show()
+--------------------+-----+--------------------+----------+
| features|label| probability|prediction|
+--------------------+-----+--------------------+----------+
|[-1.5213158492369...| 0.0|[0.99999999999278...| 0|
|[-0.2089130919726...| 1.0|[5.51104734455841...| 1|
|[-0.0776728162462...| 1.0|[6.94499110009014...| 1|
|[0.70976883811231...| 2.0|[9.24177181915992...| 2|
|[2.02217159537659...| 2.0|[2.49896507749226...| 2|
|[0.31604801093303...| 2.0|[6.91948378303032...| 2|
+--------------------+-----+--------------------+----------+
Supervised Learning¶
[68]:
train_df_scaled = train_df_scaled.select('features', 'label')
[69]:
test_df_scaled = test_df_scaled.select('features', 'label')
[70]:
from pyspark.ml.classification import RandomForestClassifier
[71]:
rf = RandomForestClassifier(
numTrees=10,
maxDepth=3,
seed=0
)
[72]:
rf = rf.fit(train_df_scaled)
[73]:
test_df_scaled = rf.transform(test_df_scaled)
[74]:
test_df_scaled.sample(0.1).show()
+--------------------+-----+--------------------+--------------------+----------+
| features|label| rawPrediction| probability|prediction|
+--------------------+-----+--------------------+--------------------+----------+
|[0.31604801093303...| 1.0|[0.03030303030303...|[0.00303030303030...| 1.0|
+--------------------+-----+--------------------+--------------------+----------+
Model evaluation¶
[75]:
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
[76]:
evaluator = MulticlassClassificationEvaluator()
[77]:
evaluator.evaluate(test_df_scaled)
[77]:
0.9618662587412588
[78]:
evaluator = evaluator.setMetricName('accuracy')
[79]:
evaluator.evaluate(test_df_scaled)
[79]:
0.9615384615384616
Pipelines¶
We can assemble pipeline for convenience and reproducibility. This is very similar to what you would do with sklearn, except that MLLib allows you to handle massive datasets by distributing the analysis to multiple computers.
We will put all the preceding steps into the pipeline, and also add hyperparameter optimization via cross-validation.
[80]:
from pyspark.ml import Pipeline
[81]:
from pyspark.ml.tuning import CrossValidator, ParamGridBuilder
[82]:
assembler = VectorAssembler(
inputCols=iris.columns[:-1],
outputCol='raw_features'
)
scaler = StandardScaler(
withMean=True,
withStd=True,
inputCol='raw_features',
outputCol='features'
)
indexer = StringIndexer(
inputCol="species",
outputCol="label"
)
rf = RandomForestClassifier()
params = (
ParamGridBuilder().
addGrid(rf.numTrees, [5,10,25]).
addGrid(rf.maxDepth, [3,4,5]).
build()
)
evaluator = MulticlassClassificationEvaluator(
metricName='accuracy'
)
cv = CrossValidator(
estimator=rf,
evaluator=evaluator,
estimatorParamMaps=params,
numFolds=3,
seed=0
).setParallelism(4)
[83]:
pipeline = Pipeline(
stages=[assembler, scaler, indexer, cv]
)
[84]:
train_iris, test_iris = iris.randomSplit([2.0, 1.0], seed=0)
[85]:
model = pipeline.fit(train_iris)
[86]:
model.stages
[86]:
[VectorAssembler_b707c6dd0daa,
StandardScalerModel: uid=StandardScaler_6f905091c141, numFeatures=4, withMean=true, withStd=true,
StringIndexerModel: uid=StringIndexer_680263d99f3c, handleInvalid=error,
CrossValidatorModel_a1e2fa6e9d8f]
[87]:
df_cv = pd.DataFrame(list(zip(model.stages[3].getEstimatorParamMaps(), model.stages[3].avgMetrics)))
df_cv[['numTrees', 'maxDepth']] = df_cv[0].apply(lambda x: pd.Series(list(x.values())))
df_cv = df_cv.drop(0, axis=1)
df_cv = df_cv.iloc[:, [1,2,0]]
df_cv.columns = df_cv.columns[:2].tolist() + ['accuracy']
df_cv.sort_values('accuracy', ascending=False)
[87]:
| numTrees | maxDepth | accuracy | |
|---|---|---|---|
| 6 | 25 | 3 | 0.958970 |
| 7 | 25 | 4 | 0.958970 |
| 8 | 25 | 5 | 0.958970 |
| 5 | 10 | 5 | 0.948217 |
| 2 | 5 | 5 | 0.941426 |
| 0 | 5 | 3 | 0.940685 |
| 3 | 10 | 3 | 0.936723 |
| 4 | 10 | 4 | 0.936723 |
| 1 | 5 | 4 | 0.929932 |
[88]:
prediction = model.transform(test_iris)
[89]:
evaluator.evaluate(prediction)
[89]:
0.9807692307692307
Using Spark for non-MLLib models¶
We can also run non-MLLIB machine learning algorithms using Spark.
Hyper-parameter optimization¶
[90]:
! python3 -m pip install --quiet joblibspark
[91]:
from sklearn.svm import SVC
from sklearn.model_selection import train_test_split, GridSearchCV
from sklearn.utils import parallel_backend
from joblibspark import register_spark
[92]:
url = 'https://bit.ly/3eoBK6t'
iris = pd.read_csv(url)
X = iris.iloc[:, :-1]
y = iris.species.astype('category').cat.codes
X_train, X_test, y_train, y_test = train_test_split(X, y)
[93]:
svc = SVC(random_state=0)
params = dict(
C = [0.1, 1, 10],
kernel = ['linear', 'poly', 'rbf', 'sigmoid']
)
cv = GridSearchCV(svc, params, cv=3)
[94]:
register_spark()
[95]:
with parallel_backend('spark', n_jobs=4):
cv.fit(X_train, y_train)
[96]:
pd.DataFrame(cv.cv_results_)
[96]:
| mean_fit_time | std_fit_time | mean_score_time | std_score_time | param_C | param_kernel | params | split0_test_score | split1_test_score | split2_test_score | mean_test_score | std_test_score | rank_test_score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.003228 | 0.000145 | 0.002065 | 0.000645 | 0.1 | linear | {'C': 0.1, 'kernel': 'linear'} | 0.973684 | 0.972973 | 0.972973 | 0.973210 | 0.000335 | 3 |
| 1 | 0.003249 | 0.000082 | 0.001860 | 0.000418 | 0.1 | poly | {'C': 0.1, 'kernel': 'poly'} | 0.973684 | 0.972973 | 0.972973 | 0.973210 | 0.000335 | 3 |
| 2 | 0.003456 | 0.000053 | 0.001981 | 0.000424 | 0.1 | rbf | {'C': 0.1, 'kernel': 'rbf'} | 0.763158 | 0.864865 | 0.756757 | 0.794927 | 0.049523 | 9 |
| 3 | 0.003865 | 0.000290 | 0.002149 | 0.000506 | 0.1 | sigmoid | {'C': 0.1, 'kernel': 'sigmoid'} | 0.368421 | 0.351351 | 0.351351 | 0.357041 | 0.008047 | 10 |
| 4 | 0.003543 | 0.000460 | 0.002302 | 0.000091 | 1 | linear | {'C': 1, 'kernel': 'linear'} | 1.000000 | 0.972973 | 0.972973 | 0.981982 | 0.012741 | 1 |
| 5 | 0.003509 | 0.000452 | 0.002630 | 0.000460 | 1 | poly | {'C': 1, 'kernel': 'poly'} | 0.947368 | 0.945946 | 0.972973 | 0.955429 | 0.012419 | 6 |
| 6 | 0.003940 | 0.000466 | 0.002706 | 0.000336 | 1 | rbf | {'C': 1, 'kernel': 'rbf'} | 0.973684 | 0.972973 | 0.972973 | 0.973210 | 0.000335 | 3 |
| 7 | 0.004211 | 0.000627 | 0.003025 | 0.000451 | 1 | sigmoid | {'C': 1, 'kernel': 'sigmoid'} | 0.368421 | 0.351351 | 0.351351 | 0.357041 | 0.008047 | 10 |
| 8 | 0.003760 | 0.000423 | 0.002659 | 0.000333 | 10 | linear | {'C': 10, 'kernel': 'linear'} | 0.947368 | 0.972973 | 0.918919 | 0.946420 | 0.022078 | 7 |
| 9 | 0.004453 | 0.000755 | 0.002508 | 0.000103 | 10 | poly | {'C': 10, 'kernel': 'poly'} | 0.921053 | 0.972973 | 0.918919 | 0.937648 | 0.024994 | 8 |
| 10 | 0.004332 | 0.000757 | 0.003176 | 0.000364 | 10 | rbf | {'C': 10, 'kernel': 'rbf'} | 1.000000 | 0.972973 | 0.972973 | 0.981982 | 0.012741 | 1 |
| 11 | 0.005562 | 0.000661 | 0.003428 | 0.000818 | 10 | sigmoid | {'C': 10, 'kernel': 'sigmoid'} | 0.210526 | 0.297297 | 0.243243 | 0.250356 | 0.035779 | 12 |
[97]:
url = 'https://bit.ly/3eoBK6t'
iris = pd.read_csv(url)
X = iris.iloc[:, :-1]
y = iris.species.astype('category').cat.codes
X_train, X_test, y_train, y_test = train_test_split(X, y)
[98]:
n, p = X_train.shape
[99]:
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import cross_val_score
[100]:
import optuna
class Objective(object):
def __init__(self, X, y):
self.X = X
self.y = y
def __call__(self, trial):
X, y = self.X, self.y # load data once only
criterion = trial.suggest_categorical('criterion', ['gini', 'entropy'])
n_estimators = trial.suggest_int('n_estimators', 50, 201)
max_features = trial.suggest_float('max_features', 1/p, 1)
max_depth = trial.suggest_float('max_depth', 1, 128, log=True)
min_samples_split = trial.suggest_int('min_samples_split', 2, 11, 1)
min_samples_leaf = trial.suggest_int('min_samples_leaf', 1, 11, 1)
clf = RandomForestClassifier(
criterion = criterion,
n_estimators = n_estimators,
max_features = max_features,
max_depth = max_depth,
min_samples_split = min_samples_split,
min_samples_leaf = min_samples_leaf,
)
score = cross_val_score(clf, X, y, cv=5).mean()
return score
objective = Objective(X_train, y_train)
study = optuna.create_study(direction='maximize', storage='sqlite:///memory')
n_jobs = 4
with parallel_backend('spark', n_jobs=n_jobs):
study.optimize(objective, n_trials=100, n_jobs=n_jobs)
[I 2020-11-04 12:47:30,511] A new study created in RDB with name: no-name-97aab8f6-8b87-4d03-afe2-5f85415b9671
[101]:
study.trials_dataframe().head(10)
[101]:
| number | value | datetime_start | datetime_complete | duration | params_criterion | params_max_depth | params_max_features | params_min_samples_leaf | params_min_samples_split | params_n_estimators | state | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 0.804348 | 2020-11-04 12:47:31.659174 | 2020-11-04 12:47:32.204716 | 0 days 00:00:00.545542 | entropy | 1.966860 | 0.457199 | 9 | 8 | 74 | COMPLETE |
| 1 | 1 | 0.964032 | 2020-11-04 12:47:31.659435 | 2020-11-04 12:47:32.745937 | 0 days 00:00:01.086502 | gini | 1.792140 | 0.997070 | 1 | 10 | 147 | COMPLETE |
| 2 | 2 | 0.955336 | 2020-11-04 12:47:31.659935 | 2020-11-04 12:47:32.810389 | 0 days 00:00:01.150454 | entropy | 118.759561 | 0.262684 | 1 | 8 | 146 | COMPLETE |
| 3 | 3 | 0.964427 | 2020-11-04 12:47:31.660439 | 2020-11-04 12:47:32.619121 | 0 days 00:00:00.958682 | gini | 4.435854 | 0.761082 | 7 | 4 | 128 | COMPLETE |
| 4 | 4 | 0.964427 | 2020-11-04 12:47:33.254434 | 2020-11-04 12:47:34.555648 | 0 days 00:00:01.301214 | entropy | 15.285156 | 0.795445 | 6 | 10 | 180 | COMPLETE |
| 5 | 5 | 0.937154 | 2020-11-04 12:47:33.691416 | 2020-11-04 12:47:34.149519 | 0 days 00:00:00.458103 | gini | 9.179145 | 0.301613 | 10 | 3 | 60 | COMPLETE |
| 6 | 6 | 0.964427 | 2020-11-04 12:47:33.780986 | 2020-11-04 12:47:34.522994 | 0 days 00:00:00.742008 | gini | 21.264760 | 0.675328 | 5 | 8 | 102 | COMPLETE |
| 7 | 7 | 0.964427 | 2020-11-04 12:47:33.831960 | 2020-11-04 12:47:34.710602 | 0 days 00:00:00.878642 | gini | 4.772650 | 0.518421 | 2 | 9 | 119 | COMPLETE |
| 8 | 8 | 0.955336 | 2020-11-04 12:47:35.174596 | 2020-11-04 12:47:36.389685 | 0 days 00:00:01.215089 | entropy | 30.773008 | 0.600859 | 8 | 5 | 175 | COMPLETE |
| 9 | 9 | 0.855336 | 2020-11-04 12:47:35.531175 | 2020-11-04 12:47:36.231259 | 0 days 00:00:00.700084 | entropy | 1.937602 | 0.955016 | 6 | 10 | 99 | COMPLETE |
Using spark with a non-MLLib classifier¶
While there is no easy way to train a general ML model in parallel, we can use Spark for distributed predictions once the model is trained,
[102]:
url = 'https://bit.ly/3eoBK6t'
iris = pd.read_csv(url)
X = iris.iloc[:, :-1]
y = iris.species.astype('category').cat.codes
X_train, X_test, y_train, y_test = train_test_split(X, y)
We use the best parameters found by Optuna in the distributed hyperparameter optimization above.
[103]:
study.best_params
[103]:
{'criterion': 'gini',
'max_depth': 4.435854141101209,
'max_features': 0.7610816811817611,
'min_samples_leaf': 7,
'min_samples_split': 4,
'n_estimators': 128}
[104]:
rf = RandomForestClassifier(**study.best_params, random_state=0)
[105]:
rf.fit(X_train, y_train)
[105]:
RandomForestClassifier(max_depth=4.435854141101209,
max_features=0.7610816811817611, min_samples_leaf=7,
min_samples_split=4, n_estimators=128, random_state=0)
[106]:
from joblib import dump, load
dump(rf, 'rf.joblib')
[106]:
['rf.joblib']
[107]:
def predict(iterartor):
model = load('rf.joblib') # model is only loaded once
for features in iterartor:
yield pd.DataFrame(model.predict(features))
[108]:
df = spark.createDataFrame(X_test)
[109]:
help(df.mapInPandas)
Help on method mapInPandas in module pyspark.sql.pandas.map_ops:
mapInPandas(func, schema) method of pyspark.sql.dataframe.DataFrame instance
Maps an iterator of batches in the current :class:`DataFrame` using a Python native
function that takes and outputs a pandas DataFrame, and returns the result as a
:class:`DataFrame`.
The function should take an iterator of `pandas.DataFrame`\s and return
another iterator of `pandas.DataFrame`\s. All columns are passed
together as an iterator of `pandas.DataFrame`\s to the function and the
returned iterator of `pandas.DataFrame`\s are combined as a :class:`DataFrame`.
Each `pandas.DataFrame` size can be controlled by
`spark.sql.execution.arrow.maxRecordsPerBatch`.
:param func: a Python native function that takes an iterator of `pandas.DataFrame`\s, and
outputs an iterator of `pandas.DataFrame`\s.
:param schema: the return type of the `func` in PySpark. The value can be either a
:class:`pyspark.sql.types.DataType` object or a DDL-formatted type string.
>>> from pyspark.sql.functions import pandas_udf
>>> df = spark.createDataFrame([(1, 21), (2, 30)], ("id", "age"))
>>> def filter_func(iterator):
... for pdf in iterator:
... yield pdf[pdf.id == 1]
>>> df.mapInPandas(filter_func, df.schema).show() # doctest: +SKIP
+---+---+
| id|age|
+---+---+
| 1| 21|
+---+---+
.. seealso:: :meth:`pyspark.sql.functions.pandas_udf`
.. note:: Experimental
.. versionadded:: 3.0
[110]:
pred = df.mapInPandas(predict, 'pred double')
[111]:
rf.predict(X_test)[:5]
[111]:
array([2, 1, 1, 1, 1], dtype=int8)
[112]:
pred.show(5)
+----+
|pred|
+----+
| 2.0|
| 1.0|
| 1.0|
| 1.0|
| 1.0|
+----+
only showing top 5 rows
[ ]: