[1]:
import warnings
warnings.simplefilter('ignore', FutureWarning)
warnings.simplefilter('ignore', UserWarning)
Imbalanced data¶
Imbalanced data occurs in classification when the number of instances in each class are not the same. Some care is required to learn to predict the rare classes effectively.
There is no one-size-fits-all approach to handling imbalanced data. A reasonable strategy is to consider this as a model selection problem, and use cross-validation to find an approach that works well for your data sets. We will show how to do this in the hyper-parameter optimization notebook.
Warning: Like most things in ML, techniques should not be applied blindly, but considered carefully with the problem goal in mind. In many cases, there is a decision-theoretic problem of assigning the appropriate costs to minority and majority case mistakes that requires domain knowledge to model correctly. As you will see in this example, blind application of a technique does not necessarily improve performance.
Simulate an imbalanced data set¶
[2]:
import pandas as pd
import numpy as np
[3]:
X_train = pd.read_csv('data/X_train.csv')
X_test = pd.read_csv('data/X_test.csv')
y_train = pd.read_csv('data/y_train.csv')
y_test = pd.read_csv('data/y_test.csv')
X = pd.concat([X_train, X_test])
y = pd.concat([y_train, y_test]).squeeze()
[4]:
y.value_counts()
[4]:
0 809
1 500
Name: survived, dtype: int64
[5]:
np.random.seed(0)
[6]:
idx = (
(y == 0) |
((y == 1) & (np.random.uniform(0, 1, y.shape) < 0.2))
).squeeze()
[7]:
X_im, y_im = X.loc[idx, :], y[idx]
[8]:
from sklearn.model_selection import train_test_split
[9]:
X_train, X_test, y_train, y_test = train_test_split(X_im, y_im, random_state=0)
[10]:
y_test.value_counts(), y_train.value_counts()
[10]:
(0 204
1 25
Name: survived, dtype: int64,
0 605
1 80
Name: survived, dtype: int64)
Collect more data¶
This is the best but often impractical solution. Synthetic data generation may also be an option.
Use evaluation metrics that are less sensitive to imbalance¶
For example, the F1 score (harmonic mean of precision and recall) is less sensitive than the accuracy score.
[11]:
from sklearn.linear_model import LogisticRegression
from sklearn.utils import class_weight
from sklearn.metrics import roc_auc_score, confusion_matrix
[12]:
from sklearn.dummy import DummyClassifier
[13]:
clf = DummyClassifier(strategy='prior')
[14]:
clf.fit(X_train, y_train)
[14]:
DummyClassifier(strategy='prior')
[15]:
clf.score(X_test, y_test)
[15]:
0.8908296943231441
[16]:
from sklearn.metrics import accuracy_score, f1_score, balanced_accuracy_score
[17]:
accuracy_score(clf.predict(X_test), y_test)
[17]:
0.8908296943231441
[18]:
f1_score(clf.predict(X_test), y_test)
[18]:
0.0
[19]:
lr = LogisticRegression()
[20]:
lr.fit(X_train, y_train)
[20]:
LogisticRegression()
[21]:
accuracy_score(lr.predict(X_test), y_test)
[21]:
0.9213973799126638
[22]:
balanced_accuracy_score(lr.predict(X_test), y_test)
[22]:
0.8723936613844872
[23]:
f1_score(lr.predict(X_test), y_test)
[23]:
0.5
Over-sample the minority class¶
There are many ways to over-sample the minority class. A popular algorithm is known as SMOTE (Synthetic Minority Oversampling Technique)
[24]:
! python3 -m pip install --quiet imbalanced-learn
[25]:
import imblearn
[26]:
X_train_resampled, y_train_resampled = \
imblearn.over_sampling.SMOTE().fit_resample(X_train, y_train)
[27]:
X_train.shape
[27]:
(685, 11)
[28]:
X_train_resampled.shape
[28]:
(1210, 11)
[29]:
y_train.value_counts()
[29]:
0 605
1 80
Name: survived, dtype: int64
Evaluate if this helps¶
[30]:
lr = LogisticRegression()
[31]:
lr.fit(X_train, y_train)
[31]:
LogisticRegression()
[32]:
f1_score(lr.predict(X_test), y_test)
[32]:
0.5
[33]:
confusion_matrix(lr.predict(X_test), y_test)
[33]:
array([[202, 16],
[ 2, 9]])
[34]:
lr.fit(X_train_resampled, y_train_resampled)
[34]:
LogisticRegression()
[35]:
f1_score(lr.predict(X_test), y_test)
[35]:
0.4109589041095891
[36]:
confusion_matrix(lr.predict(X_test), y_test)
[36]:
array([[171, 10],
[ 33, 15]])
Under-sample the majority class¶
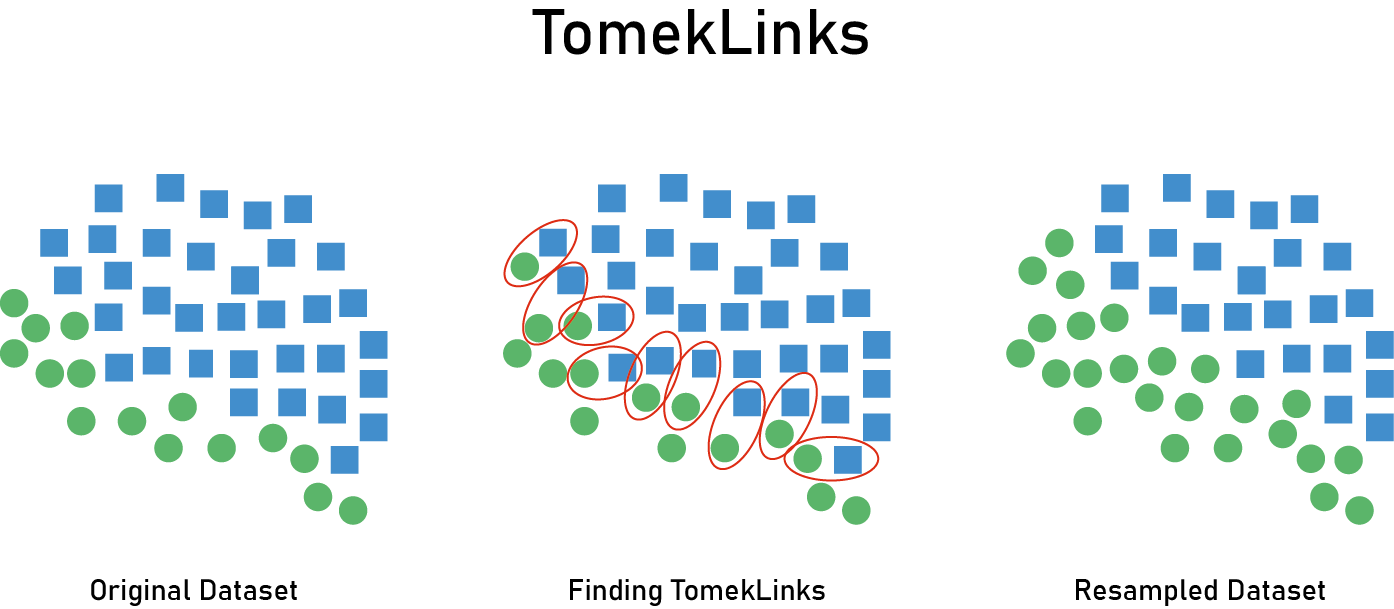
Tomek pairs are nearest neighbor pairs of instances where the classes are different. Under-sampling is done by removing the majority member of the pair.

[37]:
X_train_resampled, y_train_resampled = \
imblearn.under_sampling.TomekLinks().fit_resample(X_train, y_train)
[38]:
X_train.shape
[38]:
(685, 11)
[39]:
X_train_resampled.shape
[39]:
(665, 11)
[40]:
y_train.value_counts()
[40]:
0 605
1 80
Name: survived, dtype: int64
[41]:
y_train_resampled.value_counts()
[41]:
0 585
1 80
Name: survived, dtype: int64
Evaluate if this helps¶
[42]:
lr = LogisticRegression()
[43]:
lr.fit(X_train, y_train)
[43]:
LogisticRegression()
[44]:
f1_score(lr.predict(X_test), y_test)
[44]:
0.5
[45]:
confusion_matrix(lr.predict(X_test), y_test)
[45]:
array([[202, 16],
[ 2, 9]])
[46]:
lr.fit(X_train_resampled, y_train_resampled)
[46]:
LogisticRegression()
[47]:
f1_score(lr.predict(X_test), y_test)
[47]:
0.5
[48]:
confusion_matrix(lr.predict(X_test), y_test)
[48]:
array([[202, 16],
[ 2, 9]])
Combine over- and under-sampling¶
For example, over-sample using SMOTE then clean using Tomek.
[49]:
X_train_resampled, y_train_resampled = \
imblearn.combine.SMOTETomek().fit_resample(X_train, y_train)
[50]:
X_train.shape
[50]:
(685, 11)
[51]:
X_train_resampled.shape
[51]:
(1174, 11)
[52]:
y_train.value_counts()
[52]:
0 605
1 80
Name: survived, dtype: int64
[53]:
y_train_resampled.value_counts()
[53]:
1 587
0 587
Name: survived, dtype: int64
Evaluate if this helps¶
[54]:
lr = LogisticRegression()
[55]:
lr.fit(X_train, y_train)
[55]:
LogisticRegression()
[56]:
f1_score(lr.predict(X_test), y_test)
[56]:
0.5
[57]:
confusion_matrix(lr.predict(X_test), y_test)
[57]:
array([[202, 16],
[ 2, 9]])
[58]:
lr.fit(X_train_resampled, y_train_resampled)
[58]:
LogisticRegression()
[59]:
f1_score(lr.predict(X_test), y_test)
[59]:
0.40540540540540543
[60]:
confusion_matrix(lr.predict(X_test), y_test)
[60]:
array([[170, 10],
[ 34, 15]])
Use class weights to adjust the loss function¶
We make prediction errors in the minority class more costly than prediction errors in the majority class.
[61]:
wts = class_weight.compute_class_weight('balanced', np.unique(y_train), y_train)
[62]:
wts
[62]:
array([0.5661157, 4.28125 ])
You can then pass in the class weights. Note that there are several alternative ways to calculate possible class weights to use, and you can also do a GridSearch on weights.
This is actually built-in to most classifiers. The defaults are equal weights to each class.
[63]:
lr = LogisticRegression(class_weight=wts)
[64]:
lr.fit(X_train, y_train)
[64]:
LogisticRegression(class_weight=array([0.5661157, 4.28125 ]))
[65]:
lr.class_weight
[65]:
array([0.5661157, 4.28125 ])
[66]:
f1_score(lr.predict(X_test), y_test)
[66]:
0.5
[67]:
roc_auc_score(lr.predict(X_test), y_test)
[67]:
0.8723936613844872
[68]:
confusion_matrix(lr.predict(X_test), y_test)
[68]:
array([[202, 16],
[ 2, 9]])
[69]:
lr_balanced = LogisticRegression(class_weight='balabced')
[70]:
lr_balanced.class_weight
[70]:
'balabced'
[71]:
lr_balanced.fit(X_train, y_train)
[71]:
LogisticRegression(class_weight='balabced')
[72]:
roc_auc_score(lr_balanced.predict(X_test), y_test)
[72]:
0.8723936613844872
[73]:
confusion_matrix(lr_balanced.predict(X_test), y_test)
[73]:
array([[202, 16],
[ 2, 9]])
[74]:
f1_score(lr_balanced.predict(X_test), y_test)
[74]:
0.5
Use a classifier that is less sensitive to imbalance¶
Boosted trees are generally good because of their sequential nature.
[75]:
from catboost import CatBoostClassifier
[76]:
cb = CatBoostClassifier()
[77]:
cb.fit(X_train, y_train, verbose=0)
[77]:
<catboost.core.CatBoostClassifier at 0x135202880>
[78]:
f1_score(cb.predict(X_test), y_test)
[78]:
0.5263157894736842
[79]:
confusion_matrix(cb.predict(X_test), y_test)
[79]:
array([[201, 15],
[ 3, 10]])
Imbalanced learn has classifiers that balance the data automatically¶
[80]:
from imblearn.ensemble import BalancedRandomForestClassifier
[81]:
brf = BalancedRandomForestClassifier()
[82]:
brf.fit(X_train, y_train)
[82]:
BalancedRandomForestClassifier()
[83]:
confusion_matrix(brf.predict(X_test), y_test)
[83]:
array([[154, 8],
[ 50, 17]])
[84]:
f1_score(brf.predict(X_test), y_test)
[84]:
0.3695652173913044