Time Series Analysis II¶
In the first lecture, we are mainly concerned with how to model and evaluate time series data.
References
Statistical forecasting: notes on regression and time series analysis
Time Series Analysis (TSA) in Python - Linear Models to GARCH
Some Python packages for Time Series modeling
`tslearn<https://github.com/rtavenar/tslearn>`__`traces<https://github.com/datascopeanalytics/traces>`__`prophet<https://github.com/facebook/prophet>`__`statsmodels<https://github.com/facebook/prophet>`__`cesium<https://github.com/cesium-ml/cesium>`__`pyflux<https://github.com/RJT1990/pyflux>`__
[1]:
import warnings
warnings.simplefilter('ignore', UserWarning)
warnings.simplefilter('ignore', FutureWarning)
warnings.simplefilter('ignore', RuntimeWarning)
Stationarity¶
A stationary process is a time series whose mean, variance and auto-covariance do not change over time. Often, transformations can be applied to convert a non-stationary process to a stationary one. Periodicity (seasonality) is another form of non-stationarity that must be accounted for in the modeling.
Example¶
[2]:
import numpy as np
import pandas as pd
[3]:
df = pd.read_csv('data/uk-deaths-from-bronchitis-emphys.csv')
[4]:
df.head(3)
[4]:
| Month | U.K. deaths from bronchitis, emphysema and asthma | |
|---|---|---|
| 0 | 1974-01 | 3035 |
| 1 | 1974-02 | 2552 |
| 2 | 1974-03 | 2704 |
[5]:
df.tail(3)
[5]:
| Month | U.K. deaths from bronchitis, emphysema and asthma | |
|---|---|---|
| 70 | 1979-11 | 1781 |
| 71 | 1979-12 | 1915 |
| 72 | U.K. deaths from bronchitis | emphysema and asthma |
[6]:
df = df.iloc[:-1, :]
[7]:
df.columns = ['ds', 'y']
[8]:
df['y'] = df['y'].astype('int')
[9]:
index = pd.to_datetime(df['ds'], format='%Y-%m').copy()
[10]:
df.index = index
[11]:
df.index.freq = 'MS'
[12]:
df.drop('ds', axis=1, inplace=True)
[13]:
df.plot()
pass

Mean¶
[14]:
import matplotlib.pyplot as plt
[15]:
plt.plot(df)
plt.plot(df.rolling(window=12).mean())
pass

De-trending¶
We can make the mean stationary by subtracting the trend.
[16]:
df1 = df - df.rolling(window=12).mean()
plt.plot(df1)
plt.plot(df1.rolling(window=12).mean())
pass

Variance stabilizing transform¶
It is common to apply a simple variance stabilizing transform, especially if the variance depends on the mean.
[18]:
df2 = df.copy()
df2['y'] = np.log(df['y'])
[19]:
plt.plot(df2.rolling(window=12).var())
pass

Periodicity¶
A simple way to remove periodicity is by differencing. For intuition, consider the simple harmonic oscillator.
If we look at the second derivative, it is a constant with respect to time (and hence stationary). Differencing twice is a finite approximation to the second derivative, and achieves a similar effect of reducing oscillations.
[20]:
from scipy.integrate import odeint
[21]:
def f(x, t, ω2):
y, ydot = x
return [ydot, -ω2 * y]
[22]:
y0 = np.array([0,1])
ts = np.linspace(0, 20, 100)
ω2 = 1
xs = odeint(f, y0, ts, args=(ω2,))
[23]:
x = pd.Series(xs[:, 1]) # displacement over time
[24]:
x1 = x - x.shift()
x2 = x1 - x1.shift()
[25]:
plt.plot(x, label='displacement')
plt.plot(x1, label='velocity')
plt.plot(x2, label='acceleration')
plt.legend()
plt.tight_layout()

Auto-correlation¶
The auto-correlation function plots the Pearson correlation between a time series and a lagged version of the same time series.
[26]:
[pd.Series.corr(df.y, df.y.shift(i)) for i in range(24)][:3]
[26]:
[0.9999999999999998, 0.769695884934395, 0.40766948927949737]
For convenience there is also an autocorr function
[27]:
ac = [df.y.autocorr(i) for i in range(24)]
[28]:
ac[:3]
[28]:
[0.9999999999999998, 0.769695884934395, 0.40766948927949737]
[29]:
plt.stem(ac)
pass

[30]:
from pandas.plotting import autocorrelation_plot
[31]:
autocorrelation_plot(df.y)
pass

[32]:
from statsmodels.graphics.tsaplots import plot_acf, plot_pacf
[33]:
plot_acf(df.y, lags=24)
pass

Partial auto-correlation¶
The partial auto-correlation at lag \(k\) is a conditional correlation, and measures the correlation that remains after taking into account the correlations at lags smaller than \(k\). For an analogy, consider the regressions
\(y = \beta_0 + \beta_2 x^2\)
where \(\beta_2\) measures the dependency between \(y\) and \(x^2\)
and
\(y = \beta_0 + \beta_1 x + \beta_2 x^2\)
where \(\beta_2\) measures the dependency between \(y\) and \(x^2\) after accounting for the dependency between \(y\) and \(x\).
[34]:
plot_pacf(df.y, lags=24)
pass

Decomposing a model¶
The simplest models generally decompose the time series into one or more seasonality effects, a trend and the residuals.
[35]:
from statsmodels.tsa.seasonal import seasonal_decompose
[36]:
m1 = seasonal_decompose(df)
m1.plot()
pass

[37]:
import seaborn as sns
sns.set_context('notebook', font_scale=1.5)
[38]:
sns.distplot(m1.resid.dropna().values.squeeze())
pass

Classical models for time series¶
White noise¶
White noise refers to a time series that is independent and identically distributed (IID) with expectation equal to zero.
[39]:
def plot_ts(ts, lags=None):
fig = plt.figure(figsize = (8, 8))
ax1 = plt.subplot2grid((2, 2), (0, 0), colspan=2)
ax2 = plt.subplot2grid((2, 2), (1, 0))
ax3 = plt.subplot2grid((2, 2), (1, 1))
ax1.plot(ts)
ax1.plot(ts.rolling(window=lags).mean())
plot_acf(ts, ax=ax2, lags=lags)
plot_pacf(ts, ax=ax3, lags=lags)
plt.tight_layout()
return fig
[40]:
np.random.seed(123)
w = pd.Series(np.random.normal(0, 1, 100))
[41]:
plot_ts(w, lags=25)
pass

Random walk¶
A random walk has the following form
Note that a random walk is not stationary, since there is a time dependence.
Simulate a random walk¶
[42]:
n = 100
x = np.zeros(n)
w = np.random.normal(0, 1, n)
for t in range(n):
x[t] = x[t-1] + w[t]
x = pd.Series(x)
[43]:
plot_ts(x, lags=25)
[43]:


Effect of differencing on random walk¶
Differencing converts a random walk into a white noise process.
[44]:
x1 = x - x.shift()
plot_ts(x1.dropna(), lags=25)
[44]:


Auto-regressive models of order \(p\) AR(\(p\))¶
An AR model of order \(p\) has the following form
where \(\omega\) is a white noise term.
The time series is modeled as a linear combination of past observations.
Simulate an AR(1)¶
[45]:
np.random.seed(123)
n = 300
α = 0.6
x = np.zeros(n)
w = np.random.normal(0, 1, n)
for t in range(n):
x[t] = α*x[t-1] + w[t]
x = pd.Series(x)
[46]:
plot_ts(x, lags=25)
[46]:


Note that a reasonable estimate of \(p\) is the largest lag where the partial autocorrelation falls outside the 95% confidence interval. Here it is 1.
Fitting an AR model¶
[47]:
from statsmodels.tsa.ar_model import AR
[48]:
m2 = AR(x)
[49]:
m2.select_order(maxlag=25, ic='aic')
[49]:
1
[50]:
m2 = m2.fit(maxlag=25, ic='aic')
Compare estimated slope with true slope (=0)
[51]:
m2.params[0]
[51]:
-0.023342103477991587
Compare estimated \(\alpha\) with treu \(\alpha\) (=0.6)
[52]:
m2.params[1]
[52]:
0.6272271957584277
Simulate an AR(3) process¶
[53]:
from statsmodels.tsa.api import arma_generate_sample
[54]:
np.random.seed(123)
ar = np.array([1, -0.3, 0.4, -0.3])
ma = np.array([1, 0])
x = arma_generate_sample(ar=ar, ma=ma, nsample=100)
x = pd.Series(x)
[55]:
plot_ts(x, lags=25)
[55]:


Note that a reasonable estimate of \(p\) is the largest lag where the partial autocorrelation consistently falls outside the 95% confidence interval. Here it is 3
Fitting an AR model¶
[56]:
from statsmodels.tools.sm_exceptions import HessianInversionWarning, ConvergenceWarning
import warnings
warnings.simplefilter('ignore', HessianInversionWarning)
warnings.simplefilter('ignore', ConvergenceWarning)
[57]:
m3 = AR(x)
m3.select_order(maxlag=25, ic='aic')
[57]:
3
[58]:
m3 = m3.fit(maxlag=25, ic='aic')
Compare with true coefficients (0.3, -0.4, 0.3)
[59]:
m3.params[1:4]
[59]:
L1.y 0.280252
L2.y -0.331188
L3.y 0.473796
dtype: float64
Moving Average models MA(q)¶
A moving average model of order \(q\) is
The time series is modeled as a linear combination of past white noise terms.
[60]:
ar = np.array([1, 0])
ma = np.array([1, 0.3, 0.4, 0.7])
x = arma_generate_sample(ar=ar, ma=ma, nsample=100)
x = pd.Series(x)
[61]:
plot_ts(x, lags=25)
pass

Note that a reasonable estimate of \(q\) is the largest lag where the autocorrelation falls outside the 95% confidence interval. Here it is probably between 3 and 5.
[62]:
from statsmodels.tsa.arima_model import ARMA
[63]:
p = 0
q = 3
m4 = ARMA(x, order=(p, q))
m4 = m4.fit(maxlag=25, method='mle')
Compare with true coefficients (0.3, 0.4, 0.7)
[64]:
m4.params[1:4]
[64]:
ma.L1.y 0.294910
ma.L2.y 0.388307
ma.L3.y 0.777256
dtype: float64
[65]:
m4.summary2()
[65]:
| Model: | ARMA | BIC: | 298.3503 |
| Dependent Variable: | y | Log-Likelihood: | -137.66 |
| Date: | 2020-11-08 19:24 | Scale: | 1.0000 |
| No. Observations: | 100 | Method: | mle |
| Df Model: | 4 | Sample: | 0 |
| Df Residuals: | 96 | 0 | |
| Converged: | 1.0000 | S.D. of innovations: | 0.944 |
| No. Iterations: | 16.0000 | HQIC: | 290.596 |
| AIC: | 285.3244 |
| Coef. | Std.Err. | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| const | -0.0578 | 0.2291 | -0.2524 | 0.8007 | -0.5068 | 0.3911 |
| ma.L1.y | 0.2949 | 0.0758 | 3.8893 | 0.0001 | 0.1463 | 0.4435 |
| ma.L2.y | 0.3883 | 0.0588 | 6.6066 | 0.0000 | 0.2731 | 0.5035 |
| ma.L3.y | 0.7773 | 0.0715 | 10.8704 | 0.0000 | 0.6371 | 0.9174 |
| Real | Imaginary | Modulus | Frequency | |
|---|---|---|---|---|
| MA.1 | 0.3236 | -1.0085 | 1.0592 | -0.2006 |
| MA.2 | 0.3236 | 1.0085 | 1.0592 | 0.2006 |
| MA.3 | -1.1469 | -0.0000 | 1.1469 | -0.5000 |
ARMA(p, q)¶
As you might have suspected, we can combine the AR and MA models to get an ARMA model. The ARMA model takes the form
[66]:
np.random.seed(123)
ar = np.array([1, -0.3, 0.4, -0.3])
ma = np.array([1, 0.3, 0.4, 0.7])
x = arma_generate_sample(ar=ar, ma=ma, nsample=100)
x = pd.Series(x)
[67]:
plot_ts(x, lags=25)
pass

[68]:
p = 3
q = 3
m5 = ARMA(x, order=(p, q))
m5 = m5.fit(maxlag=25, method='mle')
[69]:
m5.summary2()
[69]:
| Model: | ARMA | BIC: | 340.6230 |
| Dependent Variable: | y | Log-Likelihood: | -151.89 |
| Date: | 2020-11-08 19:24 | Scale: | 1.0000 |
| No. Observations: | 100 | Method: | mle |
| Df Model: | 7 | Sample: | 0 |
| Df Residuals: | 93 | 0 | |
| Converged: | 1.0000 | S.D. of innovations: | 1.056 |
| No. Iterations: | 125.0000 | HQIC: | 328.217 |
| AIC: | 319.7816 |
| Coef. | Std.Err. | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| const | 0.0717 | 0.3634 | 0.1973 | 0.8436 | -0.6405 | 0.7839 |
| ar.L1.y | 0.2683 | 0.0988 | 2.7163 | 0.0066 | 0.0747 | 0.4619 |
| ar.L2.y | -0.3601 | 0.0947 | -3.8014 | 0.0001 | -0.5458 | -0.1745 |
| ar.L3.y | 0.3255 | 0.0971 | 3.3521 | 0.0008 | 0.1352 | 0.5158 |
| ma.L1.y | 0.3178 | 0.0519 | 6.1217 | 0.0000 | 0.2161 | 0.4196 |
| ma.L2.y | 0.4743 | 0.0576 | 8.2372 | 0.0000 | 0.3615 | 0.5872 |
| ma.L3.y | 0.9010 | 0.0676 | 13.3322 | 0.0000 | 0.7686 | 1.0335 |
| Real | Imaginary | Modulus | Frequency | |
|---|---|---|---|---|
| AR.1 | -0.2929 | -1.3152 | 1.3475 | -0.2849 |
| AR.2 | -0.2929 | 1.3152 | 1.3475 | 0.2849 |
| AR.3 | 1.6923 | -0.0000 | 1.6923 | -0.0000 |
| MA.1 | 0.2917 | -0.9566 | 1.0000 | -0.2029 |
| MA.2 | 0.2917 | 0.9566 | 1.0000 | 0.2029 |
| MA.3 | -1.1098 | -0.0000 | 1.1098 | -0.5000 |
We can loop through a range of orders (inspect the ACF and PACF plots) to choose the order for the AR model.
[70]:
best_aic = np.infty
for p in np.arange(5):
for q in np.arange(5):
try:
# We assume that the data has been detrended
m_ = ARMA(x, order=(p, q)).fit(method='mle', trend='nc')
aic_ = m_.aic
if aic_ < best_aic:
best_aic = aic_
best_order = (p, q)
best_m = m_
except:
pass
[71]:
best_order
[71]:
(3, 3)
ARIMA(p, d, q)¶
The ARIMA model adds differencing to convert a non-stationary model to stationarity. The parameter \(d\) is the number of differencings to perform.
[72]:
from statsmodels.tsa.arima_model import ARIMA
[73]:
best_aic = np.infty
for p in np.arange(5):
for d in np.arange(3):
for q in np.arange(5):
try:
# We assume that the data has been detrended
m_ = ARIMA(x, order=(p, d, q)).fit(method='mle', trend='nc')
aic_ = m_.aic
if aic_ < best_aic:
best_aic = aic_
best_order = (p, d, q)
best_m = m_
except:
pass
[74]:
best_order
[74]:
(3, 0, 3)
ARMA on UK disease data¶
[75]:
df.head()
[75]:
| y | |
|---|---|
| ds | |
| 1974-01-01 | 3035 |
| 1974-02-01 | 2552 |
| 1974-03-01 | 2704 |
| 1974-04-01 | 2554 |
| 1974-05-01 | 2014 |
[76]:
plot_ts(df, lags=25)
[76]:


[77]:
best_aic = np.infty
for p in np.arange(5, 10):
for q in np.arange(5, 10):
try:
m_ = ARMA(df, order=(p, q)).fit(method='mle')
aic_ = m_.aic
if aic_ < best_aic:
best_aic = aic_
best_order = (p, q)
best_m = m_
except:
pass
[78]:
best_order
[78]:
(7, 5)
Fit ARMA model¶
[79]:
m6 = ARMA(df, order=best_order)
m6 = m6.fit(maxlag=25, method='mle')
[80]:
m6.summary2()
[80]:
| Model: | ARMA | BIC: | 1063.5353 |
| Dependent Variable: | y | Log-Likelihood: | -501.83 |
| Date: | 2020-11-08 19:25 | Scale: | 1.0000 |
| No. Observations: | 72 | Method: | mle |
| Df Model: | 13 | Sample: | 01-01-1974 |
| Df Residuals: | 59 | 12-01-1979 | |
| Converged: | 1.0000 | S.D. of innovations: | 236.267 |
| No. Iterations: | 111.0000 | HQIC: | 1044.351 |
| AIC: | 1031.6620 |
| Coef. | Std.Err. | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| const | 2056.6432 | 36.6279 | 56.1496 | 0.0000 | 1984.8537 | 2128.4326 |
| ar.L1.y | 0.6118 | 0.0001 | 5513.1085 | 0.0000 | 0.6116 | 0.6120 |
| ar.L2.y | -0.1303 | nan | nan | nan | nan | nan |
| ar.L3.y | 0.1277 | nan | nan | nan | nan | nan |
| ar.L4.y | 0.0956 | nan | nan | nan | nan | nan |
| ar.L5.y | -1.0215 | 0.0000 | -46097.3269 | 0.0000 | -1.0215 | -1.0214 |
| ar.L6.y | 0.5048 | 0.0001 | 4997.4560 | 0.0000 | 0.5046 | 0.5050 |
| ar.L7.y | -0.2392 | 0.0001 | -2576.4962 | 0.0000 | -0.2394 | -0.2390 |
| ma.L1.y | -0.1347 | nan | nan | nan | nan | nan |
| ma.L2.y | -0.1679 | nan | nan | nan | nan | nan |
| ma.L3.y | -0.1693 | nan | nan | nan | nan | nan |
| ma.L4.y | -0.1386 | nan | nan | nan | nan | nan |
| ma.L5.y | 0.9967 | nan | nan | nan | nan | nan |
| Real | Imaginary | Modulus | Frequency | |
|---|---|---|---|---|
| AR.1 | -1.0010 | -0.0000 | 1.0010 | -0.5000 |
| AR.2 | -0.2989 | -0.9543 | 1.0000 | -0.2983 |
| AR.3 | -0.2989 | 0.9543 | 1.0000 | 0.2983 |
| AR.4 | 0.8681 | -0.4965 | 1.0000 | -0.0827 |
| AR.5 | 0.8681 | 0.4965 | 1.0000 | 0.0827 |
| AR.6 | 0.9866 | -1.7898 | 2.0437 | -0.1698 |
| AR.7 | 0.9866 | 1.7898 | 2.0437 | 0.1698 |
| MA.1 | -1.0001 | -0.0000 | 1.0001 | -0.5000 |
| MA.2 | -0.2979 | -0.9548 | 1.0002 | -0.2981 |
| MA.3 | -0.2979 | 0.9548 | 1.0002 | 0.2981 |
| MA.4 | 0.8675 | -0.5003 | 1.0014 | -0.0833 |
| MA.5 | 0.8675 | 0.5003 | 1.0014 | 0.0833 |
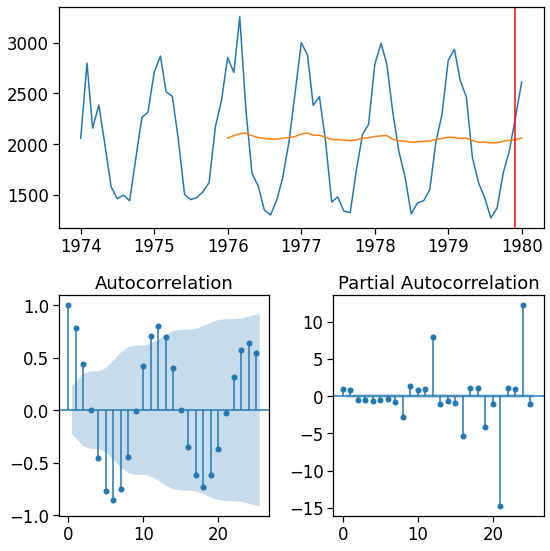
Making forecasts¶
[81]:
y_pred = m6.predict(df.index[0], df.index[-1] + pd.Timedelta(1, unit='D') )
[82]:
fig = plot_ts(y_pred, lags=25)
fig.axes[0].axvline(df.index[-1], c='red')
pass
Bayesian modeling with prophet¶
[83]:
! python3 -m pip install --quiet fbprophet
[84]:
from fbprophet import Prophet
Data needs to have just two columns ds and y.
[85]:
data = df.reset_index()
[86]:
m7 = Prophet()
[87]:
m7 = m7.fit(data)
INFO:fbprophet:Disabling weekly seasonality. Run prophet with weekly_seasonality=True to override this.
INFO:fbprophet:Disabling daily seasonality. Run prophet with daily_seasonality=True to override this.
[88]:
future = m7.make_future_dataframe(periods=24, freq='M')
[89]:
forecast = m7.predict(future)
[90]:
m7.plot(forecast)
pass

Model evaluation¶
Similarity measures¶
There are several measures commonly used to evaluate the quality of forecasts. The are the same measures we use to evaluate the fit to any function such as\(R^2\), MSE and MAE, so will not be described further here.
Cross-validation¶
From https://cdn-images-1.medium.com/max/800/1*6ujHlGolRTGvspeUDRe1EA.png

[91]:
from sklearn.model_selection import TimeSeriesSplit
from sklearn.metrics import mean_squared_error
[92]:
tsp = TimeSeriesSplit(n_splits=3)
[93]:
for train, test in tsp.split(df):
print(train.shape, test.shape)
(18,) (18,)
(36,) (18,)
(54,) (18,)
[94]:
df.index[train[-1]]
[94]:
Timestamp('1978-06-01 00:00:00', freq='MS')
[95]:
df.index[train[-1]+len(test)]
[95]:
Timestamp('1979-12-01 00:00:00', freq='MS')
A routine like the following can be used for model comparison¶
[96]:
res = []
for train, test in tsp.split(df):
m = Prophet(yearly_seasonality=False, weekly_seasonality=False, daily_seasonality=False)
m.fit(data.iloc[train])
future = data[['ds']].iloc[test]
y_pred = m.predict(future).yhat
y_true = data.y[test]
res.append(mean_squared_error(y_true, y_pred))
INFO:fbprophet:n_changepoints greater than number of observations. Using 13.
[97]:
np.mean(res)
[97]:
389145.04484811745
If you are using prophet it includees its own diagnostic functions¶
Prophet is oriented for daily data. At present, it does not appear to support cross-validation for monthly data.
