Just-in-time compilation (JIT)¶
For programmer productivity, it often makes sense to code the majority of your application in a high-level language such as Python and only optimize code bottlenecks identified by profiling. One way to speed up these bottlenecks is to compile the code to machine executables, often via an intermediate C or C-like stage. There are two common approaches to compiling Python code - using a Just-In-Time (JIT) compiler and using Cython for Ahead of Time (AOT) compilation.
This notebook mostly illustrates the JIT approach.
References
[1]:
%matplotlib inline
import matplotlib.pyplot as plt
Utility function for timing functions
We write decorators to help in timing as an alternative to timeit.
[2]:
import time
from numpy.testing import assert_almost_equal
[3]:
def timer(f, *args, **kwargs):
start = time.time()
ans = f(*args, **kwargs)
return ans, time.time() - start
[4]:
def report(fs, *args, **kwargs):
ans, t = timer(fs[0], *args, **kwargs)
print('%s: %.1f' % (fs[0].__name__, 1.0))
for f in fs[1:]:
ans_, t_ = timer(f, *args, **kwargs)
print('%s: %.1f' % (f.__name__, t/t_))
Using numexpr¶
One of the simplest approaches is to use `numexpr <https://github.com/pydata/numexpr>`__ which takes a numpy expression and compiles a more efficient version of the numpy expression written as a string. If there is a simple expression that is taking too long, this is a good choice due to its simplicity. However, it is quite limited.
[5]:
import numpy as np
a = np.random.random(int(1e6))
b = np.random.random(int(1e6))
c = np.random.random(int(1e6))
[6]:
%timeit -r3 -n3 b**2 - 4*a*c
24 ms ± 15.6 ms per loop (mean ± std. dev. of 3 runs, 3 loops each)
[7]:
import numexpr as ne
[8]:
%timeit -r3 -n3 ne.evaluate('b**2 - 4*a*c')
The slowest run took 30.97 times longer than the fastest. This could mean that an intermediate result is being cached.
12.4 ms ± 15.9 ms per loop (mean ± std. dev. of 3 runs, 3 loops each)
Using numba¶
When it works, the JIT numba can speed up Python code tremendously with minimal effort.
Documentation for ``numba` <http://numba.pydata.org/numba-doc/0.12.2/index.html>`__
Example 1¶
Plain Python version¶
[9]:
def matrix_multiply(A, B):
m, n = A.shape
n, p = B.shape
C = np.zeros((m, p))
for i in range(m):
for j in range(p):
for k in range(n):
C[i,j] += A[i,k] * B[k, j]
return C
[10]:
A = np.random.random((30, 50))
B = np.random.random((50, 40))
Numba jit version¶
[11]:
import numba
from numba import jit
[12]:
@jit
def matrix_multiply_numba(A, B):
m, n = A.shape
n, p = B.shape
C = np.zeros((m, p))
for i in range(m):
for j in range(p):
for k in range(n):
C[i,j] += A[i,k] * B[k, j]
return C
We can remove the cost of indexing a matrix in the inner loop
[13]:
@jit
def matrix_multiply_numba2(A, B):
m, n = A.shape
n, p = B.shape
C = np.zeros((m, p))
for i in range(m):
for j in range(p):
d = 0.0
for k in range(n):
d += A[i,k] * B[k, j]
C[i,j] = d
return C
[14]:
%timeit -r3 -n3 matrix_multiply(A, B)
%timeit -r3 -n3 matrix_multiply_numba(A, B)
%timeit -r3 -n3 matrix_multiply_numba2(A, B)
56.7 ms ± 787 µs per loop (mean ± std. dev. of 3 runs, 3 loops each)
The slowest run took 1842.25 times longer than the fastest. This could mean that an intermediate result is being cached.
34.5 ms ± 48.7 ms per loop (mean ± std. dev. of 3 runs, 3 loops each)
The slowest run took 1211.01 times longer than the fastest. This could mean that an intermediate result is being cached.
20.1 ms ± 28.3 ms per loop (mean ± std. dev. of 3 runs, 3 loops each)
Numpy version¶
[15]:
def matrix_multiply_numpy(A, B):
return A.dot(B)
Check that outputs are the same¶
[16]:
assert_almost_equal(matrix_multiply(A, B), matrix_multiply_numba(A, B))
assert_almost_equal(matrix_multiply(A, B), matrix_multiply_numpy(A, B))
[17]:
%timeit -r3 -n3 matrix_multiply_numba(A, B)
57.2 µs ± 3.4 µs per loop (mean ± std. dev. of 3 runs, 3 loops each)
[18]:
report([matrix_multiply, matrix_multiply_numba, matrix_multiply_numba2, matrix_multiply_numpy], A, B)
matrix_multiply: 1.0
matrix_multiply_numba: 762.8
matrix_multiply_numba2: 988.2
matrix_multiply_numpy: 111.3
Pre-compilation by giving specific signature¶
[19]:
@jit('double[:,:](double[:,:], double[:,:])')
def matrix_multiply_numba_1(A, B):
m, n = A.shape
n, p = B.shape
C = np.zeros((m, p))
for i in range(m):
for j in range(p):
d = 0.0
for k in range(n):
d += A[i,k] * B[k, j]
C[i,j] = d
return C
[20]:
%timeit -r3 -n3 matrix_multiply_numba2(A, B)
%timeit -r3 -n3 matrix_multiply_numba_1(A, B)
58.9 µs ± 2.16 µs per loop (mean ± std. dev. of 3 runs, 3 loops each)
79 µs ± 30.9 µs per loop (mean ± std. dev. of 3 runs, 3 loops each)
Example 2: Using nopython¶
Vectorized Python version¶
[21]:
def mc_pi(n):
x = np.random.uniform(-1, 1, (n,2))
return 4*np.sum((x**2).sum(1) < 1)/n
[22]:
n = int(1e6)
[23]:
mc_pi(n)
[23]:
3.141172
[24]:
%timeit mc_pi(n)
874 ms ± 12.8 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
Numba on vectorized version¶
[25]:
@jit
def mc_pi_numba(n):
x = np.random.uniform(-1, 1, (n,2))
return 4*np.sum((x**2).sum(1) < 1)/n
[26]:
%timeit mc_pi_numba(n)
39.9 ms ± 2.03 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
Using nopython¶
Using nopython, either with the @njit decorator or with @jit(nopython = True), tells numba to not use any Python objects in the C code, but only native C types. If numba cannot do this, it will raise an error. It is usually useful to run this, so you are aware of bottlenecks in your code.
[27]:
@jit(nopython=True)
def mc_pi_numba_njit(n):
x = np.random.uniform(-1, 1, (n,2))
return 4*np.sum((x**2).sum(1) < 1)/n
[28]:
%timeit mc_pi_numba_njit(n)
37.7 ms ± 2.11 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
Numba on unrolled version¶
[29]:
@jit(nopython=True)
def mc_pi_numba_unrolled(n):
s = 0
for i in range(n):
x = np.random.uniform(-1, 1)
y = np.random.uniform(-1, 1)
if (x*x + y*y) < 1:
s += 1
return 4*s/n
[30]:
mc_pi_numba_unrolled(n)
[30]:
3.143008
[31]:
%timeit -r3 -n3 mc_pi_numba_unrolled(n)
14.7 ms ± 128 µs per loop (mean ± std. dev. of 3 runs, 3 loops each)
Usig cache=True¶
This stores the compiled function in a file and avoids re-compilation on re-running a Python program.
[32]:
@jit(nopython=True, cache=True)
def mc_pi_numba_unrolled_cache(n):
s = 0
for i in range(n):
x = np.random.uniform(-1, 1)
y = np.random.uniform(-1, 1)
if (x*x + y*y) < 1:
s += 1
return 4*s/n
[33]:
%timeit -r3 -n3 mc_pi_numba_unrolled_cache(n)
16.8 ms ± 554 µs per loop (mean ± std. dev. of 3 runs, 3 loops each)
Simple parallel loops with numba¶
[34]:
from numba import njit, prange
[35]:
@njit()
def sum_rows_range(A):
s = 0
for i in range(A.shape[0]):
s += np.sum(np.exp(np.log(np.sqrt(A[i]**2.0))))
return s
[36]:
@njit(parallel=True)
def sum_rows_prange(A):
s = 0
for i in prange(A.shape[0]):
s += np.sum(np.exp(np.log(np.sqrt(A[i]**2.0))))
return s
[37]:
A = np.random.randint(0, 10, (800, 100000))
[38]:
A.shape
[38]:
(800, 100000)
Run once so that compile times excluded in benchmarking
[39]:
sum_rows_range(A), sum_rows_prange(A)
[39]:
(360006891.0, 360006891.0)
[40]:
%%time
sum_rows_range(A)
CPU times: user 5.2 s, sys: 28 ms, total: 5.22 s
Wall time: 5.18 s
[40]:
360006891.0
[41]:
%%time
sum_rows_prange(A)
CPU times: user 5.74 s, sys: 20 ms, total: 5.76 s
Wall time: 492 ms
[41]:
360006891.0
Using numba vectorize and guvectoize¶
Sometimes it is convenient to use numba to convert functions to vectorized functions for use in numpy. See documentation for details.
[42]:
from numba import int32, int64, float32, float64
Using vectorize¶
[43]:
@numba.vectorize()
def f(x, y):
return np.sqrt(x**2 + y**2)
[44]:
xs = np.random.random(10)
ys = np.random.random(10)
[45]:
np.array([np.sqrt(x**2 + y**2) for (x, y) in zip(xs, ys)])
[45]:
array([0.29016525, 0.68428663, 0.80144838, 0.94008007, 0.25911524,
0.9210368 , 0.72629006, 0.97301161, 0.42246054, 0.4597875 ])
[46]:
f(xs, ys)
[46]:
array([0.29016525, 0.68428663, 0.80144838, 0.94008007, 0.25911524,
0.9210368 , 0.72629006, 0.97301161, 0.42246054, 0.4597875 ])
Adding function signatures¶
[47]:
@numba.vectorize([float64(float64, float64),
float32(float32, float32),
float64(int64, int64),
float32(int32, int32)])
def f_sig(x, y):
return np.sqrt(x**2 + y**2)
[48]:
f_sig(xs, ys)
[48]:
array([0.29016525, 0.68428663, 0.80144838, 0.94008007, 0.25911524,
0.9210368 , 0.72629006, 0.97301161, 0.42246054, 0.4597875 ])
Using guvectorize¶
Create our own version of inner1d
Suppose we have two matrices, each with m rows. We may want to calculate an “row-wise” inner product, that is, generate a scalar for each pair of row vectors. We cannot use @vectorize because the elements are not scalars.
The layout (n),(n)->() says the function to be vectorized takes two n-element one dimensional arrays (n) and returns a scalar (). The type signature is a list that matches the order of the layout.
[49]:
@numba.guvectorize([(float64[:], float64[:], float64[:])], '(n),(n)->()')
def nb_inner1d(u, v, res):
res[0] = 0
for i in range(len(u)):
res[0] += u[i]*v[i]
[50]:
xs = np.random.random((3,4))
[51]:
nb_inner1d(xs, xs)
[51]:
array([0.57831156, 1.065156 , 1.14524983])
Check
[52]:
from numpy.core.umath_tests import inner1d
/opt/conda/lib/python3.6/site-packages/ipykernel_launcher.py:1: DeprecationWarning: numpy.core.umath_tests is an internal NumPy module and should not be imported. It will be removed in a future NumPy release.
"""Entry point for launching an IPython kernel.
[53]:
inner1d(xs,xs)
[53]:
array([0.57831156, 1.065156 , 1.14524983])
Alternative to deprecated inner1d using Einstein summation notation¶
For more on how to use Einstein notation, see the help documentation and here
[54]:
np.einsum('ij,ij->i', xs, xs)
[54]:
array([0.57831156, 1.065156 , 1.14524983])
[55]:
%timeit -r3 -n3 nb_inner1d(xs, xs)
The slowest run took 4.10 times longer than the fastest. This could mean that an intermediate result is being cached.
3.54 µs ± 2.51 µs per loop (mean ± std. dev. of 3 runs, 3 loops each)
[56]:
%timeit -r3 -n3 inner1d(xs, xs)
3.11 µs ± 2.09 µs per loop (mean ± std. dev. of 3 runs, 3 loops each)
Create our own version of matrix_multiply
[57]:
@numba.guvectorize([(int64[:,:], int64[:,:], int64[:,:])],
'(m,n),(n,p)->(m,p)')
def nb_matrix_multiply(u, v, res):
m, n = u.shape
n, p = v.shape
for i in range(m):
for j in range(p):
res[i,j] = 0
for k in range(n):
res[i,j] += u[i,k] * v[k,j]
[58]:
xs = np.random.randint(0, 10, (5, 2, 3))
ys = np.random.randint(0, 10, (5, 3, 2))
[59]:
nb_matrix_multiply(xs, ys)
[59]:
array([[[104, 62],
[ 66, 48]],
[[ 66, 26],
[ 48, 61]],
[[ 66, 35],
[ 52, 60]],
[[ 90, 85],
[ 72, 80]],
[[ 16, 46],
[ 80, 106]]])
Check
[60]:
from numpy.core.umath_tests import matrix_multiply
[61]:
matrix_multiply(xs, ys)
[61]:
array([[[104, 62],
[ 66, 48]],
[[ 66, 26],
[ 48, 61]],
[[ 66, 35],
[ 52, 60]],
[[ 90, 85],
[ 72, 80]],
[[ 16, 46],
[ 80, 106]]])
[62]:
%timeit -r3 -n3 nb_matrix_multiply(xs, ys)
The slowest run took 12.41 times longer than the fastest. This could mean that an intermediate result is being cached.
9.5 µs ± 10.5 µs per loop (mean ± std. dev. of 3 runs, 3 loops each)
[63]:
%timeit -r3 -n3 matrix_multiply(xs, ys)
3.32 µs ± 1.94 µs per loop (mean ± std. dev. of 3 runs, 3 loops each)
Parallelization with vectorize and guvectorize¶
If you have an NVidia graphics card and CUDA drivers installed, you can also use target = 'cuda'.
[64]:
@numba.vectorize([float64(float64, float64),
float32(float32, float32),
float64(int64, int64),
float32(int32, int32)],
target='parallel')
def f_parallel(x, y):
return np.sqrt(x**2 + y**2)
[65]:
xs = np.random.random(int(1e8))
ys = np.random.random(int(1e8))
[66]:
%timeit -r3 -n3 f(xs, ys)
6.01 s ± 2.25 s per loop (mean ± std. dev. of 3 runs, 3 loops each)
[67]:
%timeit -r3 -n3 f_parallel(xs, ys)
338 ms ± 32.5 ms per loop (mean ± std. dev. of 3 runs, 3 loops each)
Mandelbrot example with numba¶
Pure Python
[68]:
# color function for point at (x, y)
def mandel(x, y, max_iters):
c = complex(x, y)
z = 0.0j
for i in range(max_iters):
z = z*z + c
if z.real*z.real + z.imag*z.imag >= 4:
return i
return max_iters
[69]:
def create_fractal(xmin, xmax, ymin, ymax, image, iters):
height, width = image.shape
pixel_size_x = (xmax - xmin)/width
pixel_size_y = (ymax - ymin)/height
for x in range(width):
real = xmin + x*pixel_size_x
for y in range(height):
imag = ymin + y*pixel_size_y
color = mandel(real, imag, iters)
image[y, x] = color
[70]:
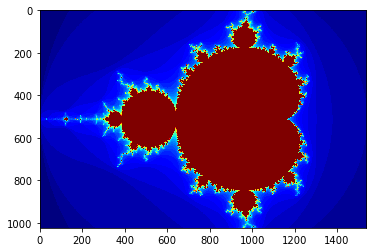
gimage = np.zeros((1024, 1536), dtype=np.uint8)
xmin, xmax, ymin, ymax = np.array([-2.0, 1.0, -1.0, 1.0]).astype('float32')
iters = 50
start = time.time()
create_fractal(xmin, xmax, ymin, ymax, gimage, iters)
dt = time.time() - start
print("Mandelbrot created on CPU in %f s" % dt)
plt.grid(False)
plt.imshow(gimage, cmap='jet')
pass
Mandelbrot created on CPU in 15.630528 s
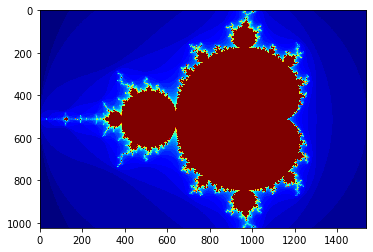
Numba
[71]:
from numba import uint32, float32
The jit decorator can also be called as a regular function
[72]:
mandel_numba = jit(uint32(float32, float32, uint32))(mandel)
[73]:
@jit
def create_fractal_numba(xmin, xmax, ymin, ymax, image, iters):
height, width = image.shape
pixel_size_x = (xmax - xmin)/width
pixel_size_y = (ymax - ymin)/height
for x in range(width):
real = xmin + x*pixel_size_x
for y in range(height):
imag = ymin + y*pixel_size_y
color = mandel_numba(real, imag, iters)
image[y, x] = color
[74]:
gimage = np.zeros((1024, 1536), dtype=np.uint8)
xmin, xmax, ymin, ymax = np.array([-2.0, 1.0, -1.0, 1.0]).astype('float32')
iters = 50
start = time.time()
create_fractal_numba(xmin, xmax, ymin, ymax, gimage, iters)
dt = time.time() - start
print("Mandelbrot created wiht Numba in %f s" % dt)
plt.grid(False)
plt.imshow(gimage, cmap='jet')
pass
Mandelbrot created wiht Numba in 0.283647 s