Tensorflow¶
[1]:
%matplotlib inline
[2]:
import warnings
warnings.simplefilter('ignore', RuntimeWarning)
[3]:
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
[4]:
from sklearn.datasets import fetch_california_housing
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
[5]:
data = fetch_california_housing()
We work with TF1 but ask it to emulate TF2 behavior
[6]:
# import tensorflow.compat.v2 as tf
# tf.enable_v2_behavior()
import tensorflow as tf
[7]:
%%capture
import tensorflow_probability as tfp
tfd = tfp.distributions
Working with tensors¶
Almost exaclty like numpy arrays.`m
[8]:
tf.constant([1., 2., 3.])
[8]:
<tf.Tensor: shape=(3,), dtype=float32, numpy=array([1., 2., 3.], dtype=float32)>
[9]:
x = tf.Variable([[1.,2.,3.], [4.,5.,6.]])
[10]:
x.shape
[10]:
TensorShape([2, 3])
[11]:
x.dtype
[11]:
tf.float32
Indexing¶
[13]:
x[:, :2]
[13]:
<tf.Tensor: shape=(2, 2), dtype=float32, numpy=
array([[1., 2.],
[4., 5.]], dtype=float32)>
Assignment¶
[14]:
x[0,:].assign([3.,2.,1.])
[14]:
<tf.Variable 'UnreadVariable' shape=(2, 3) dtype=float32, numpy=
array([[3., 2., 1.],
[4., 5., 6.]], dtype=float32)>
[15]:
x
[15]:
<tf.Variable 'Variable:0' shape=(2, 3) dtype=float32, numpy=
array([[3., 2., 1.],
[4., 5., 6.]], dtype=float32)>
Reductions¶
[16]:
tf.reduce_mean(x, axis=0)
[16]:
<tf.Tensor: shape=(3,), dtype=float32, numpy=array([3.5, 3.5, 3.5], dtype=float32)>
[17]:
tf.reduce_sum(x, axis=1)
[17]:
<tf.Tensor: shape=(2,), dtype=float32, numpy=array([ 6., 15.], dtype=float32)>
Broadcasting¶
[18]:
x + 10
[18]:
<tf.Tensor: shape=(2, 3), dtype=float32, numpy=
array([[13., 12., 11.],
[14., 15., 16.]], dtype=float32)>
[19]:
x * 10
[19]:
<tf.Tensor: shape=(2, 3), dtype=float32, numpy=
array([[30., 20., 10.],
[40., 50., 60.]], dtype=float32)>
[20]:
x - tf.reduce_mean(x, axis=1)[:, tf.newaxis]
[20]:
<tf.Tensor: shape=(2, 3), dtype=float32, numpy=
array([[ 1., 0., -1.],
[-1., 0., 1.]], dtype=float32)>
Matrix operations¶
[21]:
x @ tf.transpose(x)
[21]:
<tf.Tensor: shape=(2, 2), dtype=float32, numpy=
array([[14., 28.],
[28., 77.]], dtype=float32)>
Ufuncs¶
[22]:
tf.exp(x)
[22]:
<tf.Tensor: shape=(2, 3), dtype=float32, numpy=
array([[ 20.085537 , 7.389056 , 2.7182817],
[ 54.59815 , 148.41316 , 403.4288 ]], dtype=float32)>
[23]:
tf.sqrt(x)
[23]:
<tf.Tensor: shape=(2, 3), dtype=float32, numpy=
array([[1.7320508, 1.4142135, 1. ],
[2. , 2.236068 , 2.4494898]], dtype=float32)>
Random numbers¶
[24]:
X = tf.random.normal(shape=(10,4))
y = tf.random.normal(shape=(10,1))
Linear algebra¶
[25]:
tf.linalg.lstsq(X, y)
[25]:
<tf.Tensor: shape=(4, 1), dtype=float32, numpy=
array([[-0.427304 ],
[ 0.11494821],
[ 0.23162855],
[ 0.62853664]], dtype=float32)>
Vectorization¶
[26]:
X = tf.random.normal(shape=(1000,10,4))
y = tf.random.normal(shape=(1000,10,1))
[27]:
tf.linalg.lstsq(X, y)
[27]:
<tf.Tensor: shape=(1000, 4, 1), dtype=float32, numpy=
array([[[-7.7759638e-02],
[-1.3257186e-01],
[-7.6929688e-02],
[ 2.3807605e-01]],
[[-2.1676989e-01],
[ 1.1578191e-03],
[ 1.2898314e-01],
[ 1.9959596e-01]],
[[-5.5951335e-02],
[-2.3162486e-01],
[-1.6260512e-01],
[-2.1083586e-01]],
...,
[[ 8.7220746e-01],
[-7.2760344e-01],
[ 1.2602654e+00],
[-7.9936546e-01]],
[[ 5.4767132e-01],
[ 3.9195046e-01],
[ 4.2830217e-01],
[-4.3413453e-02]],
[[-3.0089268e-01],
[-2.1683019e-01],
[-1.4834183e-01],
[-1.8235873e-02]]], dtype=float32)>
Automatic differntiation¶
[28]:
def f(x,y):
return x**2 + 2*y**2 + 3*x*y
Gradient¶
[29]:
x, y = tf.Variable(1.0), tf.Variable(2.0)
[30]:
with tf.GradientTape() as tape:
z = f(x, y)
[31]:
tape.gradient(z, [x,y])
[31]:
[<tf.Tensor: shape=(), dtype=float32, numpy=8.0>,
<tf.Tensor: shape=(), dtype=float32, numpy=11.0>]
Hessian¶
[32]:
with tf.GradientTape(persistent=True) as H_tape:
with tf.GradientTape() as J_tape:
z = f(x, y)
Js = J_tape.gradient(z, [x,y])
Hs = [H_tape.gradient(J, [x,y]) for J in Js]
del H_tape
[33]:
np.array(Hs)
[33]:
array([[2., 3.],
[3., 4.]], dtype=float32)
Keras¶
[34]:
X_train, X_test, y_train, y_test = train_test_split(data.data, data.target)
[35]:
y_train.min(), y_train.max()
[35]:
(0.14999, 5.00001)
[36]:
scalar = StandardScaler()
X_train_s = scalar.fit_transform(X_train)
X_test_s = scalar.transform(X_test)
[37]:
import tensorflow.keras as keras
[38]:
Dense = keras.layers.Dense
We can consider a DL model as just a black box with a bunch of unnown parameters. For exanple, when the outoput is a Dense layer with just one node, the entire network model is just doing some form of regression. Hence we can replace a linear regression model with such a neural network model and run MCMC or VI as usual.
[39]:
model = keras.models.Sequential([
Dense(30,
activation='elu',
input_shape=X_train.shape[1:]),
Dense(1)
])
[40]:
model.compile(loss="mse", optimizer="nadam", metrics=["mae"])
[41]:
model.summary()
Model: "sequential"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
dense (Dense) (None, 30) 270
_________________________________________________________________
dense_1 (Dense) (None, 1) 31
=================================================================
Total params: 301
Trainable params: 301
Non-trainable params: 0
_________________________________________________________________
[42]:
model.layers
[42]:
[<tensorflow.python.keras.layers.core.Dense at 0x7f9cec625748>,
<tensorflow.python.keras.layers.core.Dense at 0x7f9cec625400>]
[43]:
model.layers[0].name
[43]:
'dense'
[44]:
model.layers[0].activation
[44]:
<function tensorflow.python.keras.activations.elu>
[45]:
hist = model.fit(X_train_s,
y_train,
epochs=10,
validation_split=0.2)
Train on 12384 samples, validate on 3096 samples
Epoch 1/10
12384/12384 [==============================] - 2s 145us/sample - loss: 1.9358 - mae: 1.0443 - val_loss: 0.6545 - val_mae: 0.5832
Epoch 2/10
12384/12384 [==============================] - 1s 60us/sample - loss: 0.5266 - mae: 0.5304 - val_loss: 0.5282 - val_mae: 0.5214
Epoch 3/10
12384/12384 [==============================] - 1s 59us/sample - loss: 0.4554 - mae: 0.4945 - val_loss: 0.4905 - val_mae: 0.5008
Epoch 4/10
12384/12384 [==============================] - 1s 60us/sample - loss: 0.4402 - mae: 0.4864 - val_loss: 0.4770 - val_mae: 0.4971
Epoch 5/10
12384/12384 [==============================] - 1s 58us/sample - loss: 0.4334 - mae: 0.4822 - val_loss: 0.4740 - val_mae: 0.4902
Epoch 6/10
12384/12384 [==============================] - 1s 58us/sample - loss: 0.4378 - mae: 0.4809 - val_loss: 0.4668 - val_mae: 0.4854
Epoch 7/10
12384/12384 [==============================] - 1s 59us/sample - loss: 0.4278 - mae: 0.4779 - val_loss: 0.4645 - val_mae: 0.4891
Epoch 8/10
12384/12384 [==============================] - 1s 59us/sample - loss: 0.4225 - mae: 0.4756 - val_loss: 0.4551 - val_mae: 0.4840
Epoch 9/10
12384/12384 [==============================] - 1s 58us/sample - loss: 0.4202 - mae: 0.4719 - val_loss: 0.4538 - val_mae: 0.4867
Epoch 10/10
12384/12384 [==============================] - 1s 58us/sample - loss: 0.4189 - mae: 0.4716 - val_loss: 0.4519 - val_mae: 0.4774
[46]:
import pandas as pd
[47]:
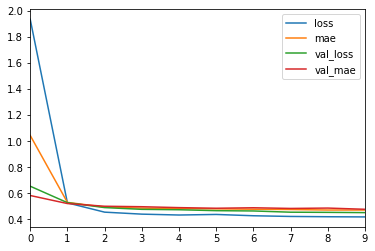
df = pd.DataFrame(hist.history)
[48]:
df.head()
[48]:
| loss | mae | val_loss | val_mae | |
|---|---|---|---|---|
| 0 | 1.935755 | 1.044255 | 0.654487 | 0.583205 |
| 1 | 0.526578 | 0.530396 | 0.528221 | 0.521408 |
| 2 | 0.455439 | 0.494459 | 0.490467 | 0.500771 |
| 3 | 0.440165 | 0.486384 | 0.476991 | 0.497115 |
| 4 | 0.433447 | 0.482155 | 0.473955 | 0.490190 |
[49]:
df.plot()
pass
[50]:
model.evaluate(X_test_s, y_test)
5160/5160 [==============================] - 0s 33us/sample - loss: 0.4417 - mae: 0.4719
[50]:
[0.4417388436868209, 0.47192544]
[51]:
np.c_[model.predict(X_test_s[:3, :]), y_test[:3]]
[51]:
array([[2.66226506, 4.889 ],
[2.38322258, 1.746 ],
[1.085361 , 1.533 ]])
[52]:
model.save('housing.h5')
[53]:
model = keras.models.load_model('housing.h5')
Tensorflow proability¶
Distributions¶
[54]:
[str(x).split('.')[-1][:-2] for x in tfd.distribution.Distribution.__subclasses__()]
[54]:
['Autoregressive',
'BatchReshape',
'Bernoulli',
'Beta',
'Categorical',
'Multinomial',
'Binomial',
'JointDistribution',
'JointDistribution',
'_Cast',
'Blockwise',
'Cauchy',
'Gamma',
'Chi2',
'TransformedDistribution',
'Normal',
'LKJ',
'CholeskyLKJ',
'_BaseDeterministic',
'_BaseDeterministic',
'Dirichlet',
'DirichletMultinomial',
'DoublesidedMaxwell',
'Empirical',
'FiniteDiscrete',
'GammaGamma',
'GaussianProcess',
'GeneralizedPareto',
'Geometric',
'Uniform',
'HalfCauchy',
'HalfNormal',
'HiddenMarkovModel',
'Horseshoe',
'Independent',
'InverseGamma',
'InverseGaussian',
'Laplace',
'LinearGaussianStateSpaceModel',
'Logistic',
'Mixture',
'MixtureSameFamily',
'MultivariateStudentTLinearOperator',
'NegativeBinomial',
'OneHotCategorical',
'Pareto',
'PERT',
'QuantizedDistribution',
'Poisson',
'_TensorCoercible',
'PixelCNN',
'PlackettLuce',
'PoissonLogNormalQuadratureCompound',
'ProbitBernoulli',
'RelaxedBernoulli',
'ExpRelaxedOneHotCategorical',
'Sample',
'StudentT',
'StudentTProcess',
'Triangular',
'TruncatedNormal',
'VectorDiffeomixture',
'VonMises',
'VonMisesFisher',
'WishartLinearOperator',
'Zipf']
[55]:
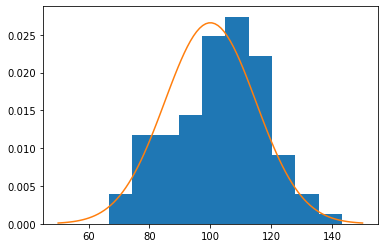
dist = tfd.Normal(loc=100, scale=15)
[56]:
x = dist.sample((3,4))
x
[56]:
<tf.Tensor: shape=(3, 4), dtype=float32, numpy=
array([[112.298294, 89.107925, 117.49498 , 105.644806],
[ 99.38011 , 121.4716 , 112.74607 , 126.83206 ],
[112.833664, 119.85916 , 105.05743 , 96.81615 ]], dtype=float32)>
[57]:
n = 100
xs = dist.sample(n)
plt.hist(xs, density=True)
xp = tf.linspace(50., 150., 100)
plt.plot(xp, dist.prob(xp))
pass
Broadcasting¶
[58]:
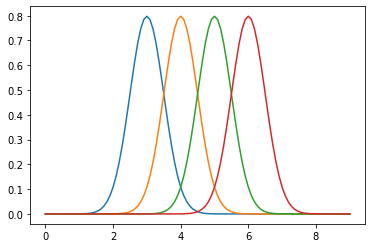
dist = tfd.Normal(loc=[3,4,5,6], scale=0.5)
[59]:
dist.sample(5)
[59]:
<tf.Tensor: shape=(5, 4), dtype=float32, numpy=
array([[3.0555534, 3.8904114, 5.2794166, 5.9625177],
[3.884369 , 5.159878 , 4.394644 , 5.358372 ],
[3.0781407, 4.152742 , 5.883699 , 5.845205 ],
[2.5866 , 3.4164045, 5.4926476, 5.5667934],
[3.1870022, 4.1119504, 5.0041285, 6.1963325]], dtype=float32)>
[60]:
xp = tf.linspace(0., 9., 100)[:, tf.newaxis]
plt.plot(np.tile(xp, dist.batch_shape), dist.prob(xp))
pass
Mixtures¶
[ ]:
tfd.MixtureSameFamily?
[61]:
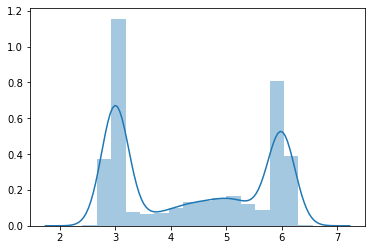
gmm = tfd.MixtureSameFamily(
mixture_distribution=tfd.Categorical(
probs=[0.4, 0.1, 0.2, 0.3]
),
components_distribution=tfd.Normal(
loc=[3., 4., 5., 6.],
scale=[0.1, 0.5, 0.5, .1])
)
[62]:
n = 10000
xs = gmm.sample(n)
[63]:
sns.distplot(xs)
pass
Transformations¶
[64]:
[x for x in dir(tfp.bijectors) if x[0].isupper()]
[64]:
['AbsoluteValue',
'Affine',
'AffineLinearOperator',
'AffineScalar',
'AutoregressiveNetwork',
'BatchNormalization',
'Bijector',
'Blockwise',
'Chain',
'CholeskyOuterProduct',
'CholeskyToInvCholesky',
'CorrelationCholesky',
'Cumsum',
'DiscreteCosineTransform',
'Exp',
'Expm1',
'FFJORD',
'FillScaleTriL',
'FillTriangular',
'GeneralizedPareto',
'Gumbel',
'GumbelCDF',
'Identity',
'Inline',
'Invert',
'IteratedSigmoidCentered',
'Kumaraswamy',
'KumaraswamyCDF',
'Log',
'Log1p',
'MaskedAutoregressiveFlow',
'MatrixInverseTriL',
'MatvecLU',
'NormalCDF',
'Ordered',
'Pad',
'Permute',
'PowerTransform',
'RationalQuadraticSpline',
'RealNVP',
'Reciprocal',
'Reshape',
'Scale',
'ScaleMatvecDiag',
'ScaleMatvecLU',
'ScaleMatvecLinearOperator',
'ScaleMatvecTriL',
'ScaleTriL',
'Shift',
'Sigmoid',
'SinhArcsinh',
'Softfloor',
'SoftmaxCentered',
'Softplus',
'Softsign',
'Square',
'Tanh',
'TransformDiagonal',
'Transpose',
'Weibull',
'WeibullCDF']
[65]:
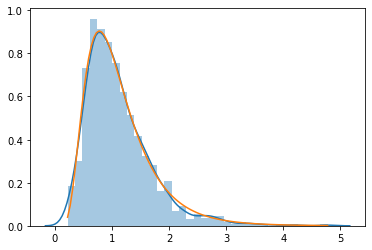
lognormal = tfp.bijectors.Exp()(tfd.Normal(0, 0.5))
[66]:
xs = lognormal.sample(1000)
sns.distplot(xs)
xp = np.linspace(tf.reduce_min(xs), tf.reduce_max(xs), 100)
plt.plot(xp, tfd.LogNormal(loc=0, scale=0.5).prob(xp))
pass
Regression¶
[67]:
xs = tf.Variable([0., 1., 2., 5., 6., 8.])
ys = tf.sin(xs) + tfd.Normal(loc=0, scale=0.5).sample(xs.shape[0])
[68]:
xs.shape, ys.shape
[68]:
(TensorShape([6]), TensorShape([6]))
[69]:
xs.numpy()
[69]:
array([0., 1., 2., 5., 6., 8.], dtype=float32)
[70]:
ys.numpy()
[70]:
array([ 0.01067163, 0.78099585, 0.6936413 , -0.37853056, 0.36542195,
0.8795101 ], dtype=float32)
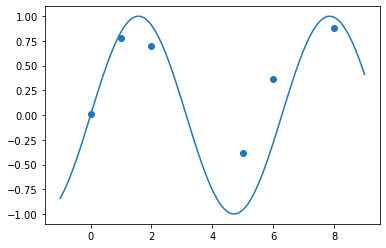
[71]:
xp = tf.linspace(-1., 9., 100)[:, None]
plt.scatter(xs.numpy(), ys.numpy())
plt.plot(xp, tf.sin(xp))
pass
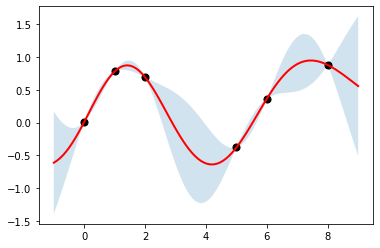
[72]:
kernel = tfp.math.psd_kernels.ExponentiatedQuadratic(length_scale=1.5)
reg = tfd.GaussianProcessRegressionModel(
kernel, xp[:, tf.newaxis], xs[:, tf.newaxis], ys
)
[74]:
ub, lb = reg.mean() + [2*reg.stddev(), -2*reg.stddev()]
plt.fill_between(np.ravel(xp), np.ravel(ub), np.ravel(lb), alpha=0.2)
plt.plot(xp, reg.mean(), c='red', linewidth=2)
plt.scatter(xs[:], ys[:], s=50, c='k')
pass
Modeling¶
Sampling from a normal distribuiton using HMC (prior predictive samples)
[75]:
[x for x in dir(tfp.mcmc) if x[0].isupper()]
[75]:
['CheckpointableStatesAndTrace',
'DualAveragingStepSizeAdaptation',
'HamiltonianMonteCarlo',
'MetropolisAdjustedLangevinAlgorithm',
'MetropolisHastings',
'NoUTurnSampler',
'RandomWalkMetropolis',
'ReplicaExchangeMC',
'SimpleStepSizeAdaptation',
'SliceSampler',
'StatesAndTrace',
'TransformedTransitionKernel',
'TransitionKernel',
'UncalibratedHamiltonianMonteCarlo',
'UncalibratedLangevin',
'UncalibratedRandomWalk']
[76]:
dir(tfp.vi)
[76]:
['__builtins__',
'__cached__',
'__doc__',
'__file__',
'__loader__',
'__name__',
'__package__',
'__path__',
'__spec__',
'_allowed_symbols',
'amari_alpha',
'arithmetic_geometric',
'chi_square',
'csiszar_vimco',
'dual_csiszar_function',
'fit_surrogate_posterior',
'jeffreys',
'jensen_shannon',
'kl_forward',
'kl_reverse',
'log1p_abs',
'modified_gan',
'monte_carlo_variational_loss',
'mutual_information',
'pearson',
'squared_hellinger',
'symmetrized_csiszar_function',
't_power',
'total_variation',
'triangular']
[77]:
from tensorflow_probability import edward2 as ed
[78]:
# From example in help docs
def unnormalized_log_prob(x):
return -x - x**2.
# Initialize the HMC transition kernel.
num_results = int(1e2)
num_burnin_steps = int(1e2)
adaptive_hmc = tfp.mcmc.SimpleStepSizeAdaptation(
tfp.mcmc.HamiltonianMonteCarlo(
target_log_prob_fn=unnormalized_log_prob,
num_leapfrog_steps=3,
step_size=1.),
num_adaptation_steps=int(num_burnin_steps * 0.8))
# Run the chain (with burn-in).
samples, is_accepted = tfp.mcmc.sample_chain(
num_results=num_results,
num_burnin_steps=num_burnin_steps,
current_state=1.,
kernel=adaptive_hmc,
trace_fn=lambda _, pkr: pkr.inner_results.is_accepted)
sample_mean = tf.reduce_mean(samples)
sample_stddev = tf.math.reduce_std(samples)
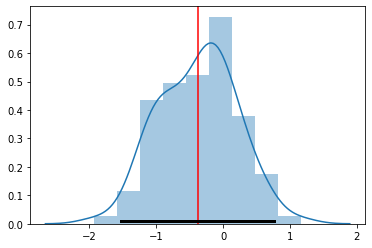
[79]:
sample_mean
[79]:
<tf.Tensor: shape=(), dtype=float32, numpy=-0.37961948>
[80]:
sample_stddev
[80]:
<tf.Tensor: shape=(), dtype=float32, numpy=0.57268214>
[81]:
sns.distplot(samples)
plt.axvline(sample_mean.numpy(), c='red')
plt.plot([sample_mean - 2*sample_stddev, sample_mean + 2*sample_stddev],
[0.01, 0.01], c='k', linewidth=3)
pass
[ ]: