[1]:
%matplotlib inline
import numpy as np
import pandas as pd
import matplotlib as mpl
import matplotlib.pyplot as plt
import seaborn as sns
import scipy.linalg as la
sns.set_context('notebook', font_scale=1.5)
1. Exact geometric solutions with \(n = m\)
Find the equation of the line that passes through the points (2,1) and (3,7)
Find the equation of the circle that passes through the points (1,7), (6,2) and (4,6)
Hint: The equation of a circle can be written as
[2]:
Find the equation of the line that passes through the points (2,1) and (3,7)
[2]:
Find the equation of the circle that passes through the points (1,7), (6,2) and (4,6)
[2]:
2. Interpolating polynomials and choice of basis
We have
\(x_i\) |
\(y_i\) |
|---|---|
0 |
-5 |
1 |
-3 |
-1 |
-15 |
2 |
39 |
-2 |
-9 |
Find interpolating polynomials using
Monomial basis \(f_i(x_i) = x^i\) - code this using only simple linear algebra operations (including solve)
Lagrange basis
\[l_j(x_j) = \prod_{0 \le m \le k, m \ne j} \frac{x - x_m}{x_j - x_m}\]
The Lagrange interpolation uses the values of \(y\) as the coefficient for the basis polynomials. Do this manually and then using the scipy.interpolate package
[2]:
x = np.array([0,1,-1,2,-2])
y = np.array([-5, -3, -15, 39, -9])
Monomial basis
[3]:
Lagrange basis
[3]:
Using library functions
[3]:
from scipy.interpolate import lagrange
[4]:
3. Markov chains
By convention, the \(rows\) of a Markov transition matrix sum to 1, and \(p_{32}\) is the probability that the system will change from state 3 to state 2. Therefore, to see the next state of an initial probability row vector \(v_k\), we need to perform left multiplication
If this is confusing, you can work with the matrix \(P^T\) and do right-multiplication with column vectors. In this case, \(p_{32}\) is the probability that the system will change from state 2 to state 3.
Find the stationary vector \(\pi^T = \pi^T P\) for the transition graph shown
by solving a set of linear equations
by solving an eigenvector problem
Check that the resulting vector is invariant with respect to the transition matrix
[4]:
Brute force check
[4]:
4. Graphs
\(M\) is the adjacency matrix of a directed graph \(G\). Find the vertices that belong to a clique.
A clique is defined as a subset of a graph where
The subset has at least 3 vertices
All pairs of vertices are connected
The subset is as large as possible
Because of the symmetry required in condition 2, we only need to consider the graph \(S\) where \(s_{ij} = 1\) if vertcies \(i\) and \(j\) communicate and 0 otherwise. Then the on-zero diagonal entries of \(S^3\) is the set states recurrent in 3 steps. That is, there is a bi-directional path \({s_i \leftrightarrow s_j \leftrightarrow s_k \leftrightarrow s_i}\), which means that the vertices \(\{s_i, s_j, s_k\}\) form a subset of a clique.
[4]:
M = np.array([
[0,1,0,1,1],
[1,0,0,1,1],
[1,1,0,1,0],
[1,1,0,0,0],
[1,0,0,1,0]
])
[5]:
5. Suppose we wish to solve the problem \(t = Mt + b\) - here the notation is from one type of such problems where \(t\) is the temperature, \(M\) is a matrix for diffusion, and \(b\) represent fixed boundary conditions. Suppose we have a 5 by 5 grid system whose boundary temperatures are fixed. Let \(M\) is a matrix with \(1/4\) for the \(\pm 1\) off-diagonals and 0 elsewhere (i.e. diffusion is approximated by the average temperature of the 4 N, S, E, W neighbors), and \(b\) is the vector \((5,2,3,3,0,1,3,0,1)\) - this assumes the temperatures along the bottom = 0, right edge = 1, top = 2 and left edge = 3. Find the equilibrium temperature at each of the 9 interior points
by solving a linear equation
by iteration
[5]:
Direct solution - not possible for large matrices
[5]:
Jacobi iteration
[5]:
6. Iterated affine maps
Define the following mapping in \(\mathbb{R}^2\)
Suppose \(s = 1/3\), \(\theta = 0\), and \(\pmatrix{a_i \\ b_i}\) are
Generate 1,000 points by first randomly selecting a point in the unit square, then applying at random one of th transformations \(T_i\) to the point. Plot the resulting 1,000 points as a scatter plot on in a square frame.
[5]:
ab =[
[0,0],
[1/3,0],
[2/3,0],
[0,1/3],
[2/3,1/3],
[0,2/3],
[1/3,2/3],
[2/3,2/3]
]
[6]:
7. The Fibonacci sequence came about from this toy model of rabbit population dynamics
A baby rabbit matures into an adult in 1 time unit
An adult gives birth to exactly 1 baby in 1 time unit
Rabbits are immortal
This gives the well known formula for the number of rabbits over discrete time \(F_{k+2} = F_{k} + F_{k+1}\)
Express this model as a matrix equation, and calculate the long-term growth rate
Let the population at any time be expreessed as the vector
In the next time step, there will be
1 adult from each adult, and one adult from each baby
1 baby from each adult
[6]:
8. Age-structured population growth
Suppose that we observe the following Leslie matrix
Starting with just 1,000 females in age-group 0-15 at time 0 and nobody else, what is the expected population after 90 years?
Suppose we could alter the fertility in a single age group for this population - can we achieve a steady state non-zero population?
[6]:
L = np.array([
[0,3,2,0.5],
[0.8,0,0,0],
[0,0.9,0,0],
[0,0,0.7,0]
])
[7]:
x0 = np.array([1000,0,0,0]).reshape(-1,1)
[8]:
Note that the real eigenvalue with real eigenvector is dominant \(\vert L_1 \vert > \vert L_k \vert\).
A theorem says this will be true if you have any two positive consecutive entries in the first row of \(L\).
The growth rate is determined by the dominant real eigenvalue with real eigenvector - in the long term, whether the population will grow, shrink or reach steady state depends on whether this is greater than, less than or equal to 1 respectively.
9.
You are given the following set of data to fit a quadratic polynomial to
x = np.arange(10)
y = np.array([ 1.58873597, 7.55101533, 10.71372171, 7.90123225,
-2.05877605, -12.40257359, -28.64568712, -46.39822281,
-68.15488905, -97.16032044])
Find the least squares solution by using the normal equations \(A^T A \hat{x} = A^T y\). (5 points)
[8]:
x = np.arange(10)
y = np.array([ 1.58873597, 7.55101533, 10.71372171, 7.90123225,
-2.05877605, -12.40257359, -28.64568712, -46.39822281,
-68.15488905, -97.16032044])
[9]:
10.
You are given the following data
A = np.array([[1, 8, 0, 7],
[0, 2, 9, 4],
[2, 8, 8, 3],
[4, 8, 6, 1],
[2, 1, 9, 6],
[0, 7, 0, 1],
[4, 0, 2, 4],
[1, 4, 9, 5],
[6, 2, 6, 6],
[9, 9, 6, 3]], dtype='float')
b = np.array([[2],
[5],
[0],
[0],
[6],
[7],
[2],
[6],
[7],
[9]], dtype='float')
Using SVD directly (not via
lstsq), find the least squares solution to \(Ax = b\) (10 points)Use SVD to find the best rank 3 approximation of A (10 points)
Calculate the approximation error in terms of the Frobenius norm (5 points)
[9]:
A = np.array([[1, 8, 0, 7],
[0, 2, 9, 4],
[2, 8, 8, 3],
[4, 8, 6, 1],
[2, 1, 9, 6],
[0, 7, 0, 1],
[4, 0, 2, 4],
[1, 4, 9, 5],
[6, 2, 6, 6],
[9, 9, 6, 3]], dtype='float')
b = np.array([[2],
[5],
[0],
[0],
[6],
[7],
[2],
[6],
[7],
[9]], dtype='float')
[10]:
11.
The page rank of a node is given by the equation
and at steady state, we have the page rank vector \(R\)
where \(d\) is the damping factor, \(N\) is the number of nodes, \(1\) is a vector of ones, and
where \(L(p_j)\) is the number of outgoing links from node \(p_j\).
Consider the graph
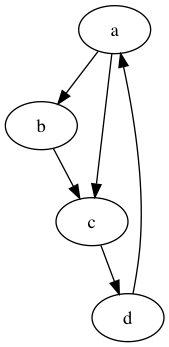
If \(d = 0.9\) find the page rank of each node
By solving a linear system
By eigendecomposition
Note: The Markov matrix constructed as instructed does not follow the usual convention. Here the columns of our Markov matrix are probability vectors, and the page rank is considered to be a column vector of the steady state probabilities.
[10]:
12.
Recall that a covariance matrix is a matrix whose entries are
Find the sample covariance matrix of the 4 features of the iris data set at http://bit.ly/2ow0oJO using basic
numpyoperations onndarrasy. Do not use thenp.covor equivalent functions inpandas(except for checking). Remember to scale by \(1/(n-1)\) for the sample covariance. (10 points)Plot the first 2 principal components of the
irisdata by using eigendecoposition, coloring each data point by the species (10 points)
[10]:
url = 'http://bit.ly/2ow0oJO'
iris = pd.read_csv(url)
iris.head()
[10]:
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
[11]:



