In [1]:
%matplotlib inline
%load_ext Cython
In [2]:
import math
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.datasets import make_blobs
from numba import jit, vectorize, float64, int64
In [3]:
sns.set_context('notebook', font_scale=1.5)
Lab10: Making Python faster¶
This homework provides practice in making Python code faster. Note that
we start with functions that already use idiomatic numpy (which are
about two orders of magnitude faster than the pure Python versions).
Functions to optimize
In [4]:
def logistic(x):
"""Logistic function."""
return np.exp(x)/(1 + np.exp(x))
def gd(X, y, beta, alpha, niter):
"""Gradient descent algorihtm."""
n, p = X.shape
Xt = X.T
for i in range(niter):
y_pred = logistic(X @ beta)
epsilon = y - y_pred
grad = Xt @ epsilon / n
beta += alpha * grad
return beta
In [5]:
x = np.linspace(-6, 6, 100)
plt.plot(x, logistic(x))
pass
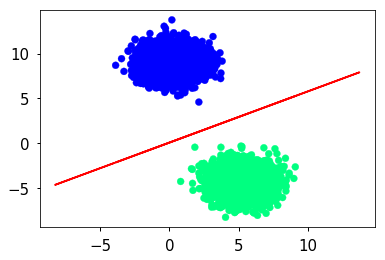
Data set for classification
In [6]:
n = 10000
p = 2
X, y = make_blobs(n_samples=n, n_features=p, centers=2, cluster_std=1.05, random_state=23)
X = np.c_[np.ones(len(X)), X]
y = y.astype('float')
Using gradient descent for classification by logistic regression
In [7]:
# initial parameters
niter = 1000
α = 0.01
β = np.zeros(p+1)
# call gradient descent
β = gd(X, y, β, α, niter)
# assign labels to points based on prediction
y_pred = logistic(X @ β)
labels = y_pred > 0.5
# calculate separating plane
sep = (-β[0] - β[1] * X)/β[2]
plt.scatter(X[:, 1], X[:, 2], c=labels, cmap='winter')
plt.plot(X, sep, 'r-')
pass
1. Rewrite the logistic function so it only makes one np.exp
call. Compare the time of both versions with the input x given below
using the @timeit magic. (10 points)
In [8]:
np.random.seed(123)
n = int(1e7)
x = np.random.normal(0, 1, n)
In [9]:
2. (20 points) Use numba to compile the gradient descent
function.
- Use the
@vectorizedecorator to create a ufunc version of the logistic function and call thislogistic_numba_cpuwith function signatures offloat64(float64). Create another function calledlogistic_numba_parallelby giving an extra argument to the decorator oftarget=parallel(5 points) - For each function, check that the answers are the same as with the
original logistic function using
np.testing.assert_array_almost_equal. Use%timeitto compare the three logistic functions (5 points) - Now use
@jitto create a JIT_compiled version of thelogisticandgdfunctions, calling themlogistic_numbaandgd_numba. Provide appropriate function signatures to the decorator in each case. (5 points) - Compare the two gradient descent functions
gdandgd_numbafor correctness and performance. (5 points)
In [12]:
3. (30 points) Use cython to compile the gradient descent
function.
- Cythonize the logistic function as
logistic_cython. Use the--annotateargument to thecythonmagic function to find slow regions. Compare accuracy and performance. The final performance should be comparable to thenumbacpu version. (10 points) - Now cythonize the gd function as
gd_cython. This function should use of the cythonizedlogistic_cythonas a C function call. Compare accuracy and performance. The final performance should be comparable to thenumbacpu version. (20 points)
Hints:
- Give static types to all variables
- Know how to use
def,cdefandcpdef - Use Typed MemoryViews
- Find out how to transpose a Typed MemoryView to store the transpose of X
- Typed MemoryVeiws are not
numpyarrays - you often have to write explicit loops to operate on them - Use the cython boundscheck, wraparound, and cdivision operators
In [ ]:
4. (40 points) Wrapping modules in C++.
Rewrite the logistic and gd functions in C++, using pybind11
to create Python wrappers. Compare accuracy and performance as usual.
Replicate the plotted example using the C++ wrapped functions for
logistic and gd
- Writing a vectorized
logisticfunction callable from both C++ and Python (10 points) - Writing the
gdfunction callable from Python (25 points) - Checking accuracy, benchmarking and creating diagnostic plots (5 points)
Hints:
- Use the C++
Eigenlibrary to do vector and matrix operations (include path is../notebooks/eigen3) - When calling the exponential function, you have to use
exp(m.array())instead ofexp(m)if you use an Eigen dynamic template. - Use
cppimportto simplify the wrapping for Python - See
`pybind11docs <http://pybind11.readthedocs.io/en/latest/index.html>`__ - See my examples for help
In [ ]: