Introduction to Scalable Data Analytics¶
We shall discuss libraries that are useful when your data is too big to fit in memory, but probably not big enough to justify the added complexity of moving to a cluster. With todays technology, this includes data sets of approximately 10s to 100s of gigabytes in size.
One goal is to introduce the storage library h5py and the pydata
packages odo, dask and blaze.
Packages to install
pip install dask
pip install cloudpickle
pip install graphviz
pip install memory_profiler
In [2]:
import time
import string
import h5py
import dask
import dask.bag as db
import dask.dataframe as dd
import dask.array as da
from dask.dot import dot_graph
from dask.imperative import do, value
from odo import odo, drop, discover
from blaze import dshape, Data, by
1 billion numbers¶
We first create 1 billion numbers in \(1000 \times 1000\) blocks and save each block in binary format.
In [71]:
def save(i, clobber=False):
fn = 'x%06d.npy' % i
if clobber or not os.path.exists(fn):
x = np.random.random((1000,1000))
np.save(fn, x)
return i
In [72]:
def save_csv(i, clobber=False):
fn = 'x%06d.csv' % i
if clobber or not os.path.exists(fn):
x = np.random.random((1000,1000))
np.savetxt(fn, x, delimiter=',', fmt='%d')
return i
In [73]:
%timeit save(0, clobber=True)
10 loops, best of 3: 63.8 ms per loop
In [74]:
%timeit save_csv(0, clobber=True)
1 loops, best of 3: 316 ms per loop
Write to disk in parallel with concurrent.futures¶
In [75]:
from concurrent.futures import ProcessPoolExecutor
In [76]:
with ProcessPoolExecutor() as pool:
pool.map(save, range(1000))
Memory usage¶
In [77]:
%load_ext memory_profiler
The memory_profiler extension is already loaded. To reload it, use:
%reload_ext memory_profiler
In [78]:
%memit x = np.load('x%06d.npy' % 0)
peak memory: 633.05 MiB, increment: 7.64 MiB
In [79]:
%memit x = np.loadtxt('x%06d.csv' % 0, delimiter=',')
peak memory: 652.47 MiB, increment: 27.05 MiB
Loading and proecessing times for a single file¶
In [80]:
def process_one(f):
x = np.load(f)
return x.mean()
In [81]:
n = 100
start = time.time()
xs =[process_one('x%06d.npy' % i) for i in range(n)]
elapsed = time.time() - start
print(np.mean(xs), 'Total: %.2fs Per file: %.2fs' % (elapsed, elapsed/n))
99.0030391889 Total: 3.31s Per file: 0.03s
Saving multiple numpy arrays to a single file¶
Using savez and savez_compressed
In [124]:
n = 100
np.savez('xs.npz', *(np.random.random((1000,1000))
for i in range(n)))
In [125]:
xs = np.load('xs.npz')
In [126]:
xs.keys()[:3]
Out[126]:
['arr_32', 'arr_19', 'arr_81']
In [127]:
xs['arr_0'].mean()
Out[127]:
0.5000693251536017
Serial code / memmap¶
Memory-mapped files are used for accessing small segments of large files
on disk, without reading the entire file into memory. The
numpy.memmap can be used anywhere an ndarray is used. The
maximum size of a memmap array is 2 GB.
In [128]:
n = 100
filename = 'random.dat'
shape = (n, 1000, 1000)
if not os.path.exists(filename):
fp = np.memmap(filename, dtype='float64', mode='w+',
shape=shape) # create memmap
for i in range(n):
x = np.load('x%06d.npy' % i)
fp[i] = x
del fp # Deletion flushes to disk before removing the object
In [129]:
fp = np.memmap(filename, dtype='float64', mode='r', shape=shape) # get handle to memmap
In [130]:
n = 100
start = time.time()
xs = [fp[i].mean() for i in range(n)]
elapsed = time.time() - start
print(np.mean(xs), 'Total: %.2fs Per file: %.2fs' % (elapsed, elapsed/n))
99.0030391889 Total: 0.62s Per file: 0.01s
Serial code / HDF5 file¶
HDF5 is a hierarchical file format that allows selective disk reads, but
also provides a tree structure for organizing your data sets. It also
does not have the size limitation of memmap and can include metadata
annotation for documentation. Because of its flexibility, you should
seriously consider using HDF5 for your data storage needs.
I suggest using the python package h5py for working wiht HDF5 files.
See documentation.
In [131]:
import datetime
In [133]:
%%time
n = 100
if not os.path.exists('random.hdf5'):
with h5py.File('random.hdf5', 'w') as f:
for i in range(n):
x = np.load('x%06d.npy' % i)
dset = f.create_dataset('x%06d' % i, shape=x.shape)
dset[:] = x
dset.attrs['created'] = str(datetime.datetime.now())
CPU times: user 898 ms, sys: 1.19 s, total: 2.09 s
Wall time: 2.24 s
In [134]:
with h5py.File('random.hdf5', 'r') as f:
f.visit(lambda x: print(x, f[x].shape, f[x].attrs['created']))
x000000 (1000, 1000) 2016-04-14 09:58:14.624532
x000001 (1000, 1000) 2016-04-14 09:58:14.644895
x000002 (1000, 1000) 2016-04-14 09:58:14.679152
x000003 (1000, 1000) 2016-04-14 09:58:14.700687
x000004 (1000, 1000) 2016-04-14 09:58:14.723214
x000005 (1000, 1000) 2016-04-14 09:58:14.747455
x000006 (1000, 1000) 2016-04-14 09:58:14.772302
x000007 (1000, 1000) 2016-04-14 09:58:14.796710
x000008 (1000, 1000) 2016-04-14 09:58:14.821258
x000009 (1000, 1000) 2016-04-14 09:58:14.842970
x000010 (1000, 1000) 2016-04-14 09:58:14.863529
x000011 (1000, 1000) 2016-04-14 09:58:14.884584
x000012 (1000, 1000) 2016-04-14 09:58:14.905112
x000013 (1000, 1000) 2016-04-14 09:58:14.928326
x000014 (1000, 1000) 2016-04-14 09:58:14.953344
x000015 (1000, 1000) 2016-04-14 09:58:14.978183
x000016 (1000, 1000) 2016-04-14 09:58:15.001180
x000017 (1000, 1000) 2016-04-14 09:58:15.021939
x000018 (1000, 1000) 2016-04-14 09:58:15.042833
x000019 (1000, 1000) 2016-04-14 09:58:15.067651
x000020 (1000, 1000) 2016-04-14 09:58:15.088946
x000021 (1000, 1000) 2016-04-14 09:58:15.109024
x000022 (1000, 1000) 2016-04-14 09:58:15.133326
x000023 (1000, 1000) 2016-04-14 09:58:15.154661
x000024 (1000, 1000) 2016-04-14 09:58:15.180672
x000025 (1000, 1000) 2016-04-14 09:58:15.202285
x000026 (1000, 1000) 2016-04-14 09:58:15.227149
x000027 (1000, 1000) 2016-04-14 09:58:15.250007
x000028 (1000, 1000) 2016-04-14 09:58:15.271642
x000029 (1000, 1000) 2016-04-14 09:58:15.292183
x000030 (1000, 1000) 2016-04-14 09:58:15.313753
x000031 (1000, 1000) 2016-04-14 09:58:15.334887
x000032 (1000, 1000) 2016-04-14 09:58:15.357399
x000033 (1000, 1000) 2016-04-14 09:58:15.378999
x000034 (1000, 1000) 2016-04-14 09:58:15.400710
x000035 (1000, 1000) 2016-04-14 09:58:15.425733
x000036 (1000, 1000) 2016-04-14 09:58:15.448780
x000037 (1000, 1000) 2016-04-14 09:58:15.470910
x000038 (1000, 1000) 2016-04-14 09:58:15.491796
x000039 (1000, 1000) 2016-04-14 09:58:15.513199
x000040 (1000, 1000) 2016-04-14 09:58:15.538417
x000041 (1000, 1000) 2016-04-14 09:58:15.559718
x000042 (1000, 1000) 2016-04-14 09:58:15.585009
x000043 (1000, 1000) 2016-04-14 09:58:15.605381
x000044 (1000, 1000) 2016-04-14 09:58:15.626484
x000045 (1000, 1000) 2016-04-14 09:58:15.647832
x000046 (1000, 1000) 2016-04-14 09:58:15.669104
x000047 (1000, 1000) 2016-04-14 09:58:15.690210
x000048 (1000, 1000) 2016-04-14 09:58:15.710586
x000049 (1000, 1000) 2016-04-14 09:58:15.731448
x000050 (1000, 1000) 2016-04-14 09:58:15.752450
x000051 (1000, 1000) 2016-04-14 09:58:15.773169
x000052 (1000, 1000) 2016-04-14 09:58:15.794386
x000053 (1000, 1000) 2016-04-14 09:58:15.819318
x000054 (1000, 1000) 2016-04-14 09:58:15.840124
x000055 (1000, 1000) 2016-04-14 09:58:15.861567
x000056 (1000, 1000) 2016-04-14 09:58:15.882513
x000057 (1000, 1000) 2016-04-14 09:58:15.906563
x000058 (1000, 1000) 2016-04-14 09:58:15.932313
x000059 (1000, 1000) 2016-04-14 09:58:15.952882
x000060 (1000, 1000) 2016-04-14 09:58:15.973459
x000061 (1000, 1000) 2016-04-14 09:58:15.994462
x000062 (1000, 1000) 2016-04-14 09:58:16.017134
x000063 (1000, 1000) 2016-04-14 09:58:16.041931
x000064 (1000, 1000) 2016-04-14 09:58:16.062847
x000065 (1000, 1000) 2016-04-14 09:58:16.083855
x000066 (1000, 1000) 2016-04-14 09:58:16.105141
x000067 (1000, 1000) 2016-04-14 09:58:16.126746
x000068 (1000, 1000) 2016-04-14 09:58:16.148961
x000069 (1000, 1000) 2016-04-14 09:58:16.174063
x000070 (1000, 1000) 2016-04-14 09:58:16.195580
x000071 (1000, 1000) 2016-04-14 09:58:16.216251
x000072 (1000, 1000) 2016-04-14 09:58:16.236949
x000073 (1000, 1000) 2016-04-14 09:58:16.259226
x000074 (1000, 1000) 2016-04-14 09:58:16.284231
x000075 (1000, 1000) 2016-04-14 09:58:16.303796
x000076 (1000, 1000) 2016-04-14 09:58:16.324429
x000077 (1000, 1000) 2016-04-14 09:58:16.349299
x000078 (1000, 1000) 2016-04-14 09:58:16.372384
x000079 (1000, 1000) 2016-04-14 09:58:16.395142
x000080 (1000, 1000) 2016-04-14 09:58:16.416727
x000081 (1000, 1000) 2016-04-14 09:58:16.436739
x000082 (1000, 1000) 2016-04-14 09:58:16.460696
x000083 (1000, 1000) 2016-04-14 09:58:16.483000
x000084 (1000, 1000) 2016-04-14 09:58:16.503930
x000085 (1000, 1000) 2016-04-14 09:58:16.525078
x000086 (1000, 1000) 2016-04-14 09:58:16.545269
x000087 (1000, 1000) 2016-04-14 09:58:16.565704
x000088 (1000, 1000) 2016-04-14 09:58:16.586277
x000089 (1000, 1000) 2016-04-14 09:58:16.608225
x000090 (1000, 1000) 2016-04-14 09:58:16.629807
x000091 (1000, 1000) 2016-04-14 09:58:16.649905
x000092 (1000, 1000) 2016-04-14 09:58:16.674557
x000093 (1000, 1000) 2016-04-14 09:58:16.695010
x000094 (1000, 1000) 2016-04-14 09:58:16.715820
x000095 (1000, 1000) 2016-04-14 09:58:16.736731
x000096 (1000, 1000) 2016-04-14 09:58:16.757302
x000097 (1000, 1000) 2016-04-14 09:58:16.778410
x000098 (1000, 1000) 2016-04-14 09:58:16.798878
x000099 (1000, 1000) 2016-04-14 09:58:16.819455
In [135]:
n = 100
start = time.time()
with h5py.File('random.hdf5', 'r') as f:
xs = [np.mean(f['x%06d' % i]) for i in range(n)]
elapsed = time.time() - start
print(np.mean(xs), 'Total: %.2fs Per file: %.2fs' % (elapsed, elapsed/n))
99.003 Total: 0.58s Per file: 0.01s
Using Dask¶
From the official documentation,
Dask is a simple task scheduling system that uses directed acyclic graphs (DAGs) of tasks to break up large computations into many small ones.
Dask enables parallel computing through task scheduling and blocked algorithms. This allows developers to write complex parallel algorithms and execute them in parallel either on a modern multi-core machine or on a distributed cluster.
On a single machine dask increases the scale of comfortable data from fits-in-memory to fits-on-disk by intelligently streaming data from disk and by leveraging all the cores of a modern CPU.
The model for how Dask works is quite similar to Spark, and we will see the same features
- lazy data structures and functions
- functional style of chaining computations and use of higher order functions
- trigger evaluations by actions
- convenience wrappers for possibly dispersed data that mimic
numpyarrays,dictsandpandasdataframes
These behave like numpy arrays, but break a massive job into
tasks that are then executed by a scheduler. The default
scheduler uses threading but you can also use multiprocessing or
distributed or even serial processing (mainly for debugging).
In [136]:
n = 100
start = time.time()
with h5py.File('random.hdf5', 'r') as f:
xs = [da.from_array(f['x%06d' % i], chunks=(1000,1000)) for i in range(n)]
xs = da.concatenate(xs)
avg = xs.mean().compute()
elapsed = time.time() - start
print(avg, 'Total: %.2fs Per file: %.2fs' % (elapsed, elapsed/n))
99.0030391889 Total: 0.96s Per file: 0.01s
Dask bags work like dictionaries for unstructured or semi-structured data sets, typically over many files.
The AA subdirectory consists of 101 1 MB plain text files from the English Wikipedia¶
In [137]:
b = db.from_filenames('data/wiki/AA/*')
In [138]:
start = time.time()
words = b.str.split().concat().frequencies().topk(10, key=lambda x: x[1])
top10 = words.compute()
elapsed = time.time() - start
print(top10, 'Total: %.2fs' % (elapsed, ))
[('the', 1051994), ('of', 617239), ('and', 482039), ('in', 370266), ('to', 356495), ('a', 312597), ('is', 174145), ('as', 145215), ('was', 141788), ('The', 141724)] Total: 90.89s
Change the scheduler¶
In [139]:
start = time.time()
words = b.str.split().concat().frequencies().topk(10, key=lambda x: x[1])
top10 = words.compute(get = dask.async.get_sync)
elapsed = time.time() - start
print(top10, 'Total: %.2fs' % (elapsed, ))
[('the', 1051994), ('of', 617239), ('and', 482039), ('in', 370266), ('to', 356495), ('a', 312597), ('is', 174145), ('as', 145215), ('was', 141788), ('The', 141724)] Total: 12.24s
Function chaining¶
In [140]:
freqs = (b.str.translate({ord(char): None for char in string.punctuation})
.str.lower()
.str.split()
.concat()
.frequencies())
In [141]:
freqs.take(5)
Out[141]:
(('statites', 2),
('tubanti', 1),
('visualisation', 8),
('manualized', 1),
('id2328', 1))
In [142]:
freqs.topk(5, key=lambda x: x[1]).compute()
Out[142]:
[('the', 1214860),
('of', 619481),
('and', 487234),
('in', 438346),
('to', 361966)]
Visualizing the task graph¶
In [143]:
dot_graph(freqs.dask)
Out[143]:
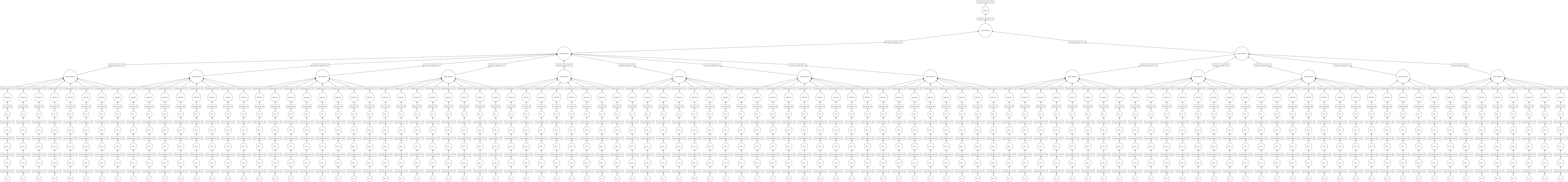
Dask dataframes¶
Dask dataframes can treat multiple pandas dataframes that might not simultaneously fit into memory like a single dataframe. See use of globbing to specify multiple source files.
In [45]:
start = time.time()
df = dd.read_csv('x00000*.csv', header=None)
print(df.describe().compute())
elapsed = time.time() - start
print(top10, 'Total: %.2fs' % (elapsed, ))
0 1 2 3 4 \
count 10000.000000 10000.000000 10000.000000 10000.000000 10000.000000
mean 99.522200 100.134400 99.460800 99.625000 100.180800
std 9.700707 10.038985 10.037736 9.917672 10.075647
min 69.000000 71.000000 62.000000 70.000000 64.000000
25% 93.000000 93.000000 93.000000 94.000000 93.000000
50% 100.000000 100.000000 100.000000 100.000000 100.000000
75% 106.000000 107.000000 106.000000 107.000000 107.000000
max 131.000000 137.000000 131.000000 137.000000 136.000000
5 6 7 8 9 \
count 10000.000000 10000.000000 10000.000000 10000.00000 10000.000000
mean 100.281200 100.254000 99.818600 99.95520 100.009600
std 10.030164 9.826736 9.794064 9.65199 10.176319
min 66.000000 64.000000 72.000000 71.00000 69.000000
25% 93.750000 94.000000 93.750000 94.00000 93.000000
50% 100.000000 100.000000 100.000000 100.00000 100.000000
75% 107.000000 107.000000 107.000000 106.25000 107.000000
max 133.000000 133.000000 136.000000 135.00000 132.000000
... 990 991 992 993 \
count ... 10000.000000 10000.000000 10000.000000 10000.000000
mean ... 99.970000 100.148800 100.098000 100.328000
std ... 10.160444 9.877126 9.931357 10.112559
min ... 67.000000 67.000000 62.000000 67.000000
25% ... 93.000000 94.000000 94.000000 93.000000
50% ... 100.000000 100.000000 100.000000 100.000000
75% ... 107.000000 108.000000 107.000000 107.000000
max ... 139.000000 131.000000 136.000000 134.000000
994 995 996 997 998 \
count 10000.000000 10000.000000 10000.000000 10000.000000 10000.000000
mean 99.636600 99.956600 99.883600 100.390200 99.722000
std 9.912475 9.999976 10.414283 9.828693 9.839959
min 70.000000 70.000000 66.000000 71.000000 67.000000
25% 93.000000 93.000000 93.000000 94.000000 93.000000
50% 100.000000 99.500000 100.000000 101.000000 100.000000
75% 106.000000 107.000000 107.000000 107.000000 107.000000
max 141.000000 133.000000 137.000000 136.000000 134.000000
999
count 10000.000000
mean 99.936400
std 9.470276
min 69.000000
25% 94.000000
50% 100.000000
75% 107.000000
max 135.000000
[8 rows x 1000 columns]
[('the', 1051994), ('of', 617239), ('and', 482039), ('in', 370266), ('to', 356495), ('a', 312597), ('is', 174145), ('as', 145215), ('was', 141788), ('The', 141724)] Total: 28.32s
Converting bags to dataframes¶
In [144]:
df_freqs = freqs.to_dataframe(columns=['word', 'n'])
df_freqs.head(10)
Out[144]:
| word | n | |
|---|---|---|
| 0 | statites | 2 |
| 1 | tubanti | 1 |
| 2 | visualisation | 8 |
| 3 | id1872 | 1 |
| 4 | id2328 | 1 |
| 5 | rolphton | 1 |
| 6 | enko | 2 |
| 7 | 400–500 | 3 |
| 8 | technique—known | 1 |
| 9 | komatiites | 1 |
Dask Imperative¶
Sometimes you need to run custom functions that don’t fit into the
array, bag or dataframe abstractions. Dask provides the imperative
module for this purpose with two decorators: do that wraps a
function and value that wraps classes. Apart from decorators and the
need to call compute for evaluation, you just write regular Python
code - yet it can take advantage of the Dask scheduling machinery. Note
that the for loop simply builds up a graph of necessary computations
- no computation is actually done until compute is called.
In [145]:
@do
def load(filename):
with open(filename) as f:
return f.read()
@do
def clean(data):
return (data
.translate({ord(char): None for char in string.punctuation})
.lower()
)
@do
def analyze(sequence_of_data):
wc = {}
for data in sequence_of_data:
words = data.split()
for word in words:
wc[word] = wc.get(word, 0) + 1
return wc
@do
def top_k(counts, k, **kwargs):
return sorted(counts.items(), reverse = True, **kwargs)[:k]
In [146]:
files = glob.glob('/Volumes/HD4/data/wiki/extracted/AA/*')[:3]
loaded = [load(i) for i in files]
cleaned = [clean(i) for i in loaded]
analyzed = analyze(cleaned)
top5 = top_k(analyzed, 5)
top5.compute()
Out[146]:
[('주판', 1), ('주산', 1), ('수판', 1), ('ㄢ', 1), ('ㄌㄨㄢ', 1)]
In [147]:
top_k(analyzed, 5, key=lambda x: x[1]).compute()
Out[147]:
[('the', 36659), ('of', 19458), ('and', 15522), ('in', 13509), ('to', 10843)]
Using Blaze¶
Blaze also works on heterogeneous data sets, and provides a high-level
consistent interface for working with data from mulitple sources. Under
the hood, blaze may make use of odo, dask and pandas.
Using blaze is very similar to usage pandas. See official
documentation.
See description at http://seanlahman.com/files/database/readme58.txt
In [148]:
import urllib.request
url = 'https://github.com/jknecht/baseball-archive-sqlite/raw/master/lahman2013.sqlite'
file_name = 'lahman2013.sqlite'
urllib.request.urlretrieve(url, file_name)
Out[148]:
('lahman2013.sqlite', <http.client.HTTPMessage at 0x173c9f6a0>)
In [150]:
db = Data('sqlite:///lahman2013.sqlite')
In [151]:
db.fields
Out[151]:
['AllstarFull',
'Appearances',
'AwardsManagers',
'AwardsPlayers',
'AwardsShareManagers',
'AwardsSharePlayers',
'Batting',
'BattingPost',
'Fielding',
'FieldingOF',
'FieldingPost',
'HallOfFame',
'Managers',
'ManagersHalf',
'Master',
'Pitching',
'PitchingPost',
'Salaries',
'Schools',
'SchoolsPlayers',
'SeriesPost',
'Teams',
'TeamsFranchises',
'TeamsHalf',
'temp']
In [154]:
db.Master.head(n=3)
Out[154]:
| playerID | birthYear | birthMonth | birthDay | birthCountry | birthState | birthCity | deathYear | deathMonth | deathDay | deathCountry | deathState | deathCity | nameFirst | nameLast | nameGiven | weight | height | bats | throws | debut | finalGame | retroID | bbrefID | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | aardsda01 | 1981 | 12 | 27 | USA | CO | Denver | NaN | NaN | NaN | None | None | None | David | Aardsma | David Allan | 205 | 75 | R | R | 1081227600000.0000000000 | 1380344400000.0000000000 | aardd001 | aardsda01 |
| 1 | aaronha01 | 1934 | 2 | 5 | USA | AL | Mobile | NaN | NaN | NaN | None | None | None | Hank | Aaron | Henry Louis | 180 | 72 | R | R | -496087200000.0000000000 | 213166800000.0000000000 | aaroh101 | aaronha01 |
| 2 | aaronto01 | 1939 | 8 | 5 | USA | AL | Mobile | 1984 | 8 | 16 | USA | GA | Atlanta | Tommie | Aaron | Tommie Lee | 190 | 75 | R | R | -243885600000.0000000000 | 54709200000.0000000000 | aarot101 | aaronto01 |
In [155]:
master = db.Master
birth = by(master.birthCountry, n=master.birthCountry.count())
birth.sort('n', ascending=False).head(5)
Out[155]:
| birthCountry | n | |
|---|---|---|
| 0 | USA | 16153 |
| 1 | D.R. | 592 |
| 2 | Venezuela | 301 |
| 3 | CAN | 243 |
| 4 | P.R. | 238 |